Passage
Fluorescent amino acids are useful for labeling proteins because they can be visualized spectroscopically, facilitating the study of protein structure and interactions. The noncanonical amino acid dansylalanine, a fluorescent amino acid that is sensitive to the hydrophobicity of its environment, is a good candidate to be incorporated into proteins because of its small size and fluorescent capabilities. Dansyl chloride and Compound 1 were used to synthesize dansylalanine, as shown in Reaction 1. Dansylalanine absorbs maximally at 340 nm and emits at approximately 540 nm.
 Reaction 1 Synthesis of dansylalanineA leucyl-tRNA synthetase from Escherichia coli was engineered to aminoacylate bacterial tRNAs containing a CUA anticodon with dansylalanine. After verifying that the tRNA/synthetase pair does not interact with endogenous amino acids or tRNAs, it was transfected into the yeast Saccharomyces cerevisiae along with a plasmid encoding a mutated human superoxide dismutase (hSOD) . Using this system, dansylalanine was genetically incorporated into hSOD in place of either Gln-16 or Trp-33, and expressed in S. cerevisiae.Position 16 is located on the surface of hSOD at the N-terminus of a β-strand, and position 33 lies in the center of a strand within a β-barrel. To probe changes in the structure of hSOD, researchers exposed the purified protein to guanidinium chloride (GdmCl) to denature the protein at concentrations ranging 0-4.5 M. The experiment was conducted in a 35 mM sodium phosphate buffer at pH 7.2. The fluorescence of hSOD with dansylalanine at either position 16 or 33 was monitored at each GdmCl concentration, and the results of the unfolding experiment are shown in Figure 1.
Reaction 1 Synthesis of dansylalanineA leucyl-tRNA synthetase from Escherichia coli was engineered to aminoacylate bacterial tRNAs containing a CUA anticodon with dansylalanine. After verifying that the tRNA/synthetase pair does not interact with endogenous amino acids or tRNAs, it was transfected into the yeast Saccharomyces cerevisiae along with a plasmid encoding a mutated human superoxide dismutase (hSOD) . Using this system, dansylalanine was genetically incorporated into hSOD in place of either Gln-16 or Trp-33, and expressed in S. cerevisiae.Position 16 is located on the surface of hSOD at the N-terminus of a β-strand, and position 33 lies in the center of a strand within a β-barrel. To probe changes in the structure of hSOD, researchers exposed the purified protein to guanidinium chloride (GdmCl) to denature the protein at concentrations ranging 0-4.5 M. The experiment was conducted in a 35 mM sodium phosphate buffer at pH 7.2. The fluorescence of hSOD with dansylalanine at either position 16 or 33 was monitored at each GdmCl concentration, and the results of the unfolding experiment are shown in Figure 1.
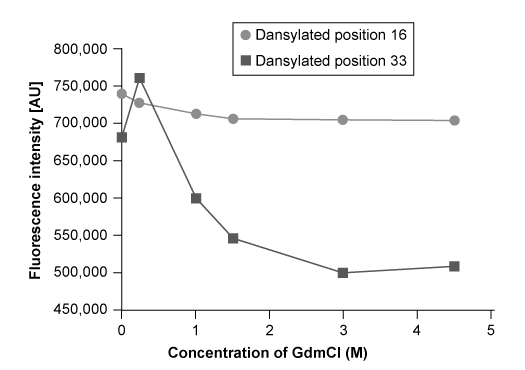 Figure 1 Plot of dansylalanine fluorescence at different concentrations of GdmCl
Figure 1 Plot of dansylalanine fluorescence at different concentrations of GdmCl
Adapted from Summerer D, Chen S, Wu N, Deiters A, Chin JW, Schultz PG. A genetically encoded fluorescent amino acid. Proc Natl Acad Sci USA. 2006.
-What is the charge of the N-terminus during the protein unfolding experiment?
A) +1, because the amine group is not protonated
B) +1, because the amine group is in the conjugate acid form
C) 0, because the amine group is protonated
D) 0, because the amine group is in the conjugate base form
Correct Answer:
Verified
Q58: Which structure is the product of the
Q59: Passage
The β-lactam scaffold is an important feature
Q60: Passage
Two key ingredients found in many soaps
Q61: If an alcohol were to undergo a
Q62: What is the correct systematic name for
Q64: Passage
Fluorescent amino acids are useful for labeling
Q65: Which of the following does NOT describe
Q66: A student needs to separate a mixture
Q67: When the compound shown below undergoes an
Q68: Two separate reactions are conducted in which
Unlock this Answer For Free Now!
View this answer and more for free by performing one of the following actions

Scan the QR code to install the App and get 2 free unlocks

Unlock quizzes for free by uploading documents