Deck 31: Completing the Protein Life Cycle: Folding, Processing, and Degradation
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/24
Play
Full screen (f)
Deck 31: Completing the Protein Life Cycle: Folding, Processing, and Degradation
1
How Do Newly Synthesized Proteins Fold?
About 40 years ago, Christian Anfinsen produced a set of experiments that proved that most proteins fold spontaneously. The specific information and interactions found in the primary structure was all that was needed for a protein to assume the correct shape.
In an intracellular environment, where many reactions are occurring simultaneously, a protein has a chance of interacting with an undesired molecule. For this reason, many proteins have acquired the help of chaperones to help produce the correct form.
Hsp70 chaperones bind to hydrophobic regions of a protein and prevent other molecules from binding. They also prevent the protein from folding until it is in the correct environment to form its correct shape. Hsp70 chaperones are ATP dependent.
Hsp60 chaperones provided an encased area or "Anfinsen cage" for the protein to produce its correct form, free from side interactions. Hsp60 chaperones have an upper limit to the size of protein that they can encase. It is also ATP dependent.
In an intracellular environment, where many reactions are occurring simultaneously, a protein has a chance of interacting with an undesired molecule. For this reason, many proteins have acquired the help of chaperones to help produce the correct form.
Hsp70 chaperones bind to hydrophobic regions of a protein and prevent other molecules from binding. They also prevent the protein from folding until it is in the correct environment to form its correct shape. Hsp70 chaperones are ATP dependent.
Hsp60 chaperones provided an encased area or "Anfinsen cage" for the protein to produce its correct form, free from side interactions. Hsp60 chaperones have an upper limit to the size of protein that they can encase. It is also ATP dependent.
2
Assessing the ATP cost of protein synthesis
(Integrates with Chapter 30.) Human rhodanese (33kD) consists of 296 amino acid residues. Approximately how many ATP equivalents are consumed in the synthesis of the rhodanese polypeptide chain from its constituent amino acids and the folding of this chain into an active tertiary structure?
(Integrates with Chapter 30.) Human rhodanese (33kD) consists of 296 amino acid residues. Approximately how many ATP equivalents are consumed in the synthesis of the rhodanese polypeptide chain from its constituent amino acids and the folding of this chain into an active tertiary structure?
Elongation of an amino acid requires 4 ATP per amino acid. The first step in elongating a polypeptide is activating the amino acid and then attaching it to a tRNA. This process converts an ATP to an AMP, costing 2 phosphate groups. This is equivalent to hydrolyzing 2 ATPs in this step.
Next the "loaded" aminoacyl-tRNA is moved to the A-site of a ribosome. This is accomplished with the help of the elongation factor EF-Tu and the hydrolysis of a GTP. The hydrolysis of GTP is energetically equivalent to hydrolyzing another ATP.
Finally, the aminoacyl-tRNA is moved from the A-site to the P-site using an EF-G:GTP complex. The GTP is again hydrolyzed to GDP. This once again is equivalent to the hydrolysis of one ATP. This leaves us with a total of 4 ATP equivalents per amino acid.
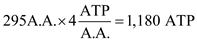 We now must consider initiation and termination. Initiation requires 2 ATP. One is in the form of GTP, for Met-tRNA i binding to form the 40S pre-initiation complex, and another is in the form of ATP in forming the initiation complex. The formation of Met-tRNA i costs another 2 ATP.
We now must consider initiation and termination. Initiation requires 2 ATP. One is in the form of GTP, for Met-tRNA i binding to form the 40S pre-initiation complex, and another is in the form of ATP in forming the initiation complex. The formation of Met-tRNA i costs another 2 ATP.
Termination requires the hydrolysis of one GTP. This means that for an amino acid that is 296 amino acids long, 4 ATP equivalents will be consumed in initiation, 1,180 ATP equivalents will be consumed in elongation and 1 ATP equivalent will be consumed in termination. This gives a total of 1,185 ATP equivalents.
Next the "loaded" aminoacyl-tRNA is moved to the A-site of a ribosome. This is accomplished with the help of the elongation factor EF-Tu and the hydrolysis of a GTP. The hydrolysis of GTP is energetically equivalent to hydrolyzing another ATP.
Finally, the aminoacyl-tRNA is moved from the A-site to the P-site using an EF-G:GTP complex. The GTP is again hydrolyzed to GDP. This once again is equivalent to the hydrolysis of one ATP. This leaves us with a total of 4 ATP equivalents per amino acid.
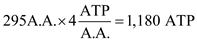 We now must consider initiation and termination. Initiation requires 2 ATP. One is in the form of GTP, for Met-tRNA i binding to form the 40S pre-initiation complex, and another is in the form of ATP in forming the initiation complex. The formation of Met-tRNA i costs another 2 ATP.
We now must consider initiation and termination. Initiation requires 2 ATP. One is in the form of GTP, for Met-tRNA i binding to form the 40S pre-initiation complex, and another is in the form of ATP in forming the initiation complex. The formation of Met-tRNA i costs another 2 ATP.Termination requires the hydrolysis of one GTP. This means that for an amino acid that is 296 amino acids long, 4 ATP equivalents will be consumed in initiation, 1,180 ATP equivalents will be consumed in elongation and 1 ATP equivalent will be consumed in termination. This gives a total of 1,185 ATP equivalents.
3
How Are Proteins Processed Following Translation?
Due to the significant number of proteins in comparison to the relatively small number of human genes, alterations outside of mRNA splicing must be available. Newly synthesized proteins almost always undergo posttranslational modifications. Many proteins can be made from a single mRNA through alternative splicing.
The N-terminal methionine residue is often proteolyzed and removed. Most newly synthesized proteins will also undergo phosphorylation, glycosylation, or another form of covalent modification.
The N-terminal methionine residue is often proteolyzed and removed. Most newly synthesized proteins will also undergo phosphorylation, glycosylation, or another form of covalent modification.
4
Understanding the consequences of proteolysis
A single proteolytic break in a polypeptide chain of a native protein is often sufficient to initiate its total degradation. What does this fact suggest to you regarding the structural consequences of proteolytic nicks in proteins?
A single proteolytic break in a polypeptide chain of a native protein is often sufficient to initiate its total degradation. What does this fact suggest to you regarding the structural consequences of proteolytic nicks in proteins?

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
5
How Do Proteins Find Their Proper Place in the Cell?

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
6
Understanding the relevance of chaperones in protein folding
Protein molecules, like all molecules, can be characterized in terms of general properties such as size, shape, charge, solubility/hydrophobicity. Consider the influence of each of these general features on the likelihood of whether folding of a particular protein will require chaperone assistance or not. Be specific regarding Hsp70 chaperones or Hsp70 chaperones and Hsp60 chaperonins.
Protein molecules, like all molecules, can be characterized in terms of general properties such as size, shape, charge, solubility/hydrophobicity. Consider the influence of each of these general features on the likelihood of whether folding of a particular protein will require chaperone assistance or not. Be specific regarding Hsp70 chaperones or Hsp70 chaperones and Hsp60 chaperonins.

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
7
How Does Protein Degradation Regulate Cellular Levels of Specific Proteins?

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
8
Assessing the chaperone-assisted folding of multi-subunit proteins
Many multidomain proteins apparently do not require chaperones to attain the fully folded conformations. Suggest a rational scenario for chaperone-independent folding of such proteins.
Many multidomain proteins apparently do not require chaperones to attain the fully folded conformations. Suggest a rational scenario for chaperone-independent folding of such proteins.

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
9
Assessing the dimensions and capacity of GroEL
The GroEL ring has a 5-nm central cavity. Calculate the maximum molecular weight for a spherical protein that can just fit into this cavity, assuming the density of the protein is 1.25 g/mL.
The GroEL ring has a 5-nm central cavity. Calculate the maximum molecular weight for a spherical protein that can just fit into this cavity, assuming the density of the protein is 1.25 g/mL.

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
10
Understanding the possible phosphorylated states of acetyl-CoA carboxylase
(Integrates with Chapter 24.) Acetyl-CoA carboxylase has at least seven possible phosphorylation sites (residues 23, 25, 29, 76, 77, 95 and 1200) in its 2345-residue polypeptide (see Figure 24.4). How many different covalently modified forms of acetyl-CoA carboxylase protein are possible if there are seven phosphorylation sites?
(Integrates with Chapter 24.) Acetyl-CoA carboxylase has at least seven possible phosphorylation sites (residues 23, 25, 29, 76, 77, 95 and 1200) in its 2345-residue polypeptide (see Figure 24.4). How many different covalently modified forms of acetyl-CoA carboxylase protein are possible if there are seven phosphorylation sites?

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
11
Comparing the mechanisms of action of EF-Tu/EF-Ts and DnaK/GrpE
(Integrates with Chapter 30.) In what ways are the mechanisms of action of EF-Tu/EF-Ts and DnaK/GrpE similar? What mechanistic functions do the ribosome A-site and DnaJ have in common?
(Integrates with Chapter 30.) In what ways are the mechanisms of action of EF-Tu/EF-Ts and DnaK/GrpE similar? What mechanistic functions do the ribosome A-site and DnaJ have in common?

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
12
Understanding the structure and function of signal sequences and signal peptidases
The amino acid sequence deduced from the nucleotide sequence of a newly discovered human gene begins: MRSLLILVLCFLPAALGK…. Is this a signal sequence? If so, where does the signal peptidase act on it? What can you surmise about the intended destination of this protein?
The amino acid sequence deduced from the nucleotide sequence of a newly discovered human gene begins: MRSLLILVLCFLPAALGK…. Is this a signal sequence? If so, where does the signal peptidase act on it? What can you surmise about the intended destination of this protein?

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
13
Understanding the mechanism of transport of integral membrane proteins
Not only is the Sec61p translocon complex essential for translocation of proteins into the ER lumen, it also mediates the incorporation of integral membrane proteins into the ER membrane. The mechanism for integration is triggered by stop-transfer signals that cause a pause in translocation. Figure 31.5 shows the translocon as a closed cylinder spanning the membrane. Suggest a mechanism for lateral transfer of an integral membrane protein from the protein-conducting channel of the translocon into the hydrophobic phase of the ER membrane.
Not only is the Sec61p translocon complex essential for translocation of proteins into the ER lumen, it also mediates the incorporation of integral membrane proteins into the ER membrane. The mechanism for integration is triggered by stop-transfer signals that cause a pause in translocation. Figure 31.5 shows the translocon as a closed cylinder spanning the membrane. Suggest a mechanism for lateral transfer of an integral membrane protein from the protein-conducting channel of the translocon into the hydrophobic phase of the ER membrane.

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
14
Assessing the dimensions and capacity of Sec61p
The Sec61p core complex of the translocon has a highly dynamic pore whose internal diameter varies from 0.6 to 6 nm. In post-translational translocation, folded proteins can move across the ER membrane through this pore. What is the molecular weight of a spherical protein that would just fit through a 6-nm pore? (Adopt the same assumptions used in problem 5.)
The Sec61p core complex of the translocon has a highly dynamic pore whose internal diameter varies from 0.6 to 6 nm. In post-translational translocation, folded proteins can move across the ER membrane through this pore. What is the molecular weight of a spherical protein that would just fit through a 6-nm pore? (Adopt the same assumptions used in problem 5.)

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
15
Assessing the dimensions and capacity of the protein conducting translocon
(Integrates with Chapters 6, 9, and 30.) During co-translational translocation, the peptide tunnel running from the peptidyl transferase center of the large ribosomal subunit and the protein conducting-channel are aligned. If the tunnel through the ribosomal subunit is 10 nm and the translocon channel has the same length as the thickness of a phospholipid bilayer, what is the minimum number of amino acid residues sequestered in this common conduit?
(Integrates with Chapters 6, 9, and 30.) During co-translational translocation, the peptide tunnel running from the peptidyl transferase center of the large ribosomal subunit and the protein conducting-channel are aligned. If the tunnel through the ribosomal subunit is 10 nm and the translocon channel has the same length as the thickness of a phospholipid bilayer, what is the minimum number of amino acid residues sequestered in this common conduit?

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
16
Understanding the nature of the ubiquitin-ubiquitin linkage
Draw the structure of the isopeptide bond formed between Gly76 of one ubiquitin molecule and Lys48 of another ubiquitin molecule.
Draw the structure of the isopeptide bond formed between Gly76 of one ubiquitin molecule and Lys48 of another ubiquitin molecule.

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
17
Understanding the nature of E3 ubiquitin protein ligation
Assign the 20 amino acids to either of two groups based on their susceptibility to ubiquitin ligation by E3 ubiquitin protein ligase. Can you discern any common attributes among the amino acids in the less susceptible versus the more susceptible group?
Assign the 20 amino acids to either of two groups based on their susceptibility to ubiquitin ligation by E3 ubiquitin protein ligase. Can you discern any common attributes among the amino acids in the less susceptible versus the more susceptible group?

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
18
Assessing the consequences of proteasome inhibition
Lactacystin is a Streptomyces natural product that acts as an irreversible inhibitor of 26S proteosome ?-subunit catalytic activity by covalent attachment to N-terminal threonine -OH groups. Predict the effects of lactacystin on cell cycle progression.
Lactacystin is a Streptomyces natural product that acts as an irreversible inhibitor of 26S proteosome ?-subunit catalytic activity by covalent attachment to N-terminal threonine -OH groups. Predict the effects of lactacystin on cell cycle progression.

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
19
Understanding the dual functionality of the HtrA protease
HtrA proteases are dual-function chaperone-protease protein quality control systems. The protease activity of HtrA proteases depends on a proper spatial relationship between the Asp-His-Ser catalytic triad. Propose a mechanism for the temperature-induced switch of HtrA proteases from chaperone function to protease function.
HtrA proteases are dual-function chaperone-protease protein quality control systems. The protease activity of HtrA proteases depends on a proper spatial relationship between the Asp-His-Ser catalytic triad. Propose a mechanism for the temperature-induced switch of HtrA proteases from chaperone function to protease function.

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
20
Understanding the scope and diversity of posttranslational protein modification
As described in this chapter, the most common post-translational modifications of proteins are proteolysis, phosphorylation, methylation, acetylation, and linkage with ubiquitin and SUMO proteins. Carry out a Web search to identify at least eight other posttranslational modifications and the amino acid residues involved in these modifications.
As described in this chapter, the most common post-translational modifications of proteins are proteolysis, phosphorylation, methylation, acetylation, and linkage with ubiquitin and SUMO proteins. Carry out a Web search to identify at least eight other posttranslational modifications and the amino acid residues involved in these modifications.

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
21
Understanding the nature and functional utility of fluorescence resonance energy transfer (FRET)
Fluorescence resonance energy transfer (FRET) is a spectroscopic technique that can be used to provide certain details of the conformation of biomolecules. Look up FRET on the Web or in an introductory text on FRET uses in biochemistry, and explain how FRET could be used to observe conformational changes in proteins bound to chaperonins such as GroEL. A good article on FRET in protein folding and dynamics can be found here: Haas, E., 2005. The study of protein folding and dynamics by determination of intramolecular distance distributions and their fluctuations using ensemble and single-molecule FRET measurements. ChemPhysChem 6:858-870. Studies of GroEL using FRET analysis include the following: Sharma, S., et al., 2008. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell 133:142-153; and Lin, Z., et al., 2008. GroEL stimulates protein folding through forced unfolding. Nature Structural and Molecular Biology 15:303-311.
Fluorescence resonance energy transfer (FRET) is a spectroscopic technique that can be used to provide certain details of the conformation of biomolecules. Look up FRET on the Web or in an introductory text on FRET uses in biochemistry, and explain how FRET could be used to observe conformational changes in proteins bound to chaperonins such as GroEL. A good article on FRET in protein folding and dynamics can be found here: Haas, E., 2005. The study of protein folding and dynamics by determination of intramolecular distance distributions and their fluctuations using ensemble and single-molecule FRET measurements. ChemPhysChem 6:858-870. Studies of GroEL using FRET analysis include the following: Sharma, S., et al., 2008. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell 133:142-153; and Lin, Z., et al., 2008. GroEL stimulates protein folding through forced unfolding. Nature Structural and Molecular Biology 15:303-311.

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
22
Understanding the nature and functions of the phosphodegron The cross-talk between phosphorylation and ubiquitination in protein degradation processes is encapsulated in the concept of the "phosphodegron." What is a phosphodegron, and how does phosphorylation serve as a recognition signal for protein degradation? (A good reference on the phosphodegron and crosstalk between phosphorylation and ubiquitination is Hunter, T., 2007. The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Molecular Cell 28:730-738.)

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
23
Assessing the consequences of a common post-translational modification
A common post-translational modification is the removal of the universal N-terminal methionine in many proteins by Met-aminopeptidase. How might Met removal affect the half-life of the protein?
A common post-translational modification is the removal of the universal N-terminal methionine in many proteins by Met-aminopeptidase. How might Met removal affect the half-life of the protein?

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck
24
Designing a sequence for a peptide with amphipathic ?-helical secondary structure
Figure 31.6 shows the generalized structure of an amphipathic ?-helix found as an N-terminal presequence on a nuclear-encoded mitochondrial protein. Write out a 20-residue-long amino acid sequence that would give rise to such an amphipathic ? -helical secondary structure.
Figure 31.6 shows the generalized structure of an amphipathic ?-helix found as an N-terminal presequence on a nuclear-encoded mitochondrial protein. Write out a 20-residue-long amino acid sequence that would give rise to such an amphipathic ? -helical secondary structure.

Unlock Deck
Unlock for access to all 24 flashcards in this deck.
Unlock Deck
k this deck



