Deck 5: The Structure of RNA
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/15
Play
Full screen (f)
Deck 5: The Structure of RNA
1
Justify the biological reason for the presence of uracil in RNA but not in DNA.
The biological reason for the presence of uracil in RNA but not in DNA has been explained below:
Adenine is present in both RNA and DNA. Adenine can pair with both uracil and thymine. Both uracil and cytosine differ in a single group only. In DNA, if spontaneous deamination occurs, it causes the change of cytosine to uracil.
If replication occurs in this strand where deamination of cytosine has taken place, the GC pair will be changed to GU pair and then finally AT pair.
If uracil is found naturally in DNA, the mismatch repair system cannot find the mutation. This would result in accumulation of mutations in DNA. Since DNA is the genetic material, mutations should not occur. RNA on the other hand is synthesized and destroyed within a few hours. So even if mutations occur in RNA, it will be destroyed and not transferred to the next generation.
Adenine is present in both RNA and DNA. Adenine can pair with both uracil and thymine. Both uracil and cytosine differ in a single group only. In DNA, if spontaneous deamination occurs, it causes the change of cytosine to uracil.
If replication occurs in this strand where deamination of cytosine has taken place, the GC pair will be changed to GU pair and then finally AT pair.
If uracil is found naturally in DNA, the mismatch repair system cannot find the mutation. This would result in accumulation of mutations in DNA. Since DNA is the genetic material, mutations should not occur. RNA on the other hand is synthesized and destroyed within a few hours. So even if mutations occur in RNA, it will be destroyed and not transferred to the next generation.
2
Given the following sequence of RNA, propose the potential hairpin structure for this RNA. Indicate base pairing with a dotted line.
5?-AGGACCCUUCGGGGUUCU-3?
5?-AGGACCCUUCGGGGUUCU-3?
The following ribonucleic acid (RNA) sequence is given:
5'- AGGACCCUUCGGGGUUCU-3'
The potential hairpin structure for this sequence would be:
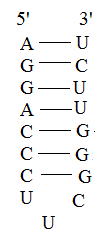 Most of the bases are complementary while three do not form any bonds. AU base pairing is normally seen while GU base pairing also occurs. CU base pairing does not occur and so the last three bases form the loop of the structure and do not form any bonds between them.
Most of the bases are complementary while three do not form any bonds. AU base pairing is normally seen while GU base pairing also occurs. CU base pairing does not occur and so the last three bases form the loop of the structure and do not form any bonds between them.
5'- AGGACCCUUCGGGGUUCU-3'
The potential hairpin structure for this sequence would be:
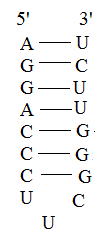 Most of the bases are complementary while three do not form any bonds. AU base pairing is normally seen while GU base pairing also occurs. CU base pairing does not occur and so the last three bases form the loop of the structure and do not form any bonds between them.
Most of the bases are complementary while three do not form any bonds. AU base pairing is normally seen while GU base pairing also occurs. CU base pairing does not occur and so the last three bases form the loop of the structure and do not form any bonds between them. 3
The yeast alanine tRNA is shown below (adapted from Chapter 2, Fig. 2-14). Identify the type(s) of secondary structure present in this tRNA. Identify any noncanonical base pairing.
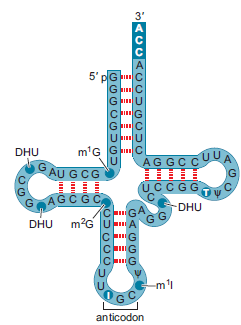

In the figure in the question, the yeast alanine transfer ribonucleic acid (tRNA) is shown. The types of secondary structures present in the tRNA are:
• Stem structure
• Loop structure
• Hairpin structure (comprising of stem and loop)The stem structure is formed between the bases found at the 3' and 5' ends. There are 3 loops seen in the structure. A small loop is also present in between two loops.
The noncanonical base pairings seen are:
• G-U
• U-U
• Stem structure
• Loop structure
• Hairpin structure (comprising of stem and loop)The stem structure is formed between the bases found at the 3' and 5' ends. There are 3 loops seen in the structure. A small loop is also present in between two loops.
The noncanonical base pairings seen are:
• G-U
• U-U
4
Researchers want to find a way to confer antibiotic resistance to selected bacterial cells. To do so, they decide to identify a sequence of RNA (aptamer) by SELEX that specifically binds to the antibiotic of interest and expresses that RNA aptamer in the selected cells to confer antibiotic resistance.
A. Given what you know about RNA, hypothesize what general properties of the RNA aptamer allow for specific binding to the antibiotic.
B. Instead of using SELEX, the researchers could start with an isolated pool of RNAs encoded by the bacterial genome and select for the RNA(s) that bind the antibiotic to identify their desired RNA. Give two advantages of using SELEX rather than this method.
A. Given what you know about RNA, hypothesize what general properties of the RNA aptamer allow for specific binding to the antibiotic.
B. Instead of using SELEX, the researchers could start with an isolated pool of RNAs encoded by the bacterial genome and select for the RNA(s) that bind the antibiotic to identify their desired RNA. Give two advantages of using SELEX rather than this method.

Unlock Deck
Unlock for access to all 15 flashcards in this deck.
Unlock Deck
k this deck
5
Describe the properties shared between an enzyme and a ribozyme.

Unlock Deck
Unlock for access to all 15 flashcards in this deck.
Unlock Deck
k this deck
6
Some RNAs act as plant pathogens or viroids. Some viroids are capable of carrying out a catalytic reaction. You are given a sequence of RNA isolated as a viroid to a specific plant species. Identify some characteristics that would indicate that this viroid acts as a catalytic hammerhead.

Unlock Deck
Unlock for access to all 15 flashcards in this deck.
Unlock Deck
k this deck
7
Scientists design modified versions of the self-cleaving hammerhead as potential therapeutics. To target cleavage of another RNA molecule, describe how the engineered structure of the hammerhead is modified. Why does this change make the hammerhead a true ribozyme?

Unlock Deck
Unlock for access to all 15 flashcards in this deck.
Unlock Deck
k this deck
8
Some ribozymes like those found in the hepatitis delta virus and the ribozyme/riboswitch found in the glmS mRNA are capable of folding into a nested double pseudoknot. Discuss several advantages to pseudoknot formation in ribozymes and riboswitches.

Unlock Deck
Unlock for access to all 15 flashcards in this deck.
Unlock Deck
k this deck
9
The structure given below is a nucleoside analog.
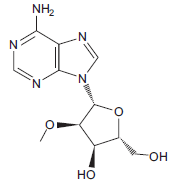
A. Name the nucleoside that the structure mimics.
B. Imagine a strand of RNA containing this nucleoside analog (linked by phosphodiester bonds as in normal RNA). In a high-pH environment, is the RNA strand subject to hydrolysis where the nucleoside analog is present? Explain why or why not.

A. Name the nucleoside that the structure mimics.
B. Imagine a strand of RNA containing this nucleoside analog (linked by phosphodiester bonds as in normal RNA). In a high-pH environment, is the RNA strand subject to hydrolysis where the nucleoside analog is present? Explain why or why not.

Unlock Deck
Unlock for access to all 15 flashcards in this deck.
Unlock Deck
k this deck
10
DNA serves as the genetic material. Give three cellular roles for RNA.

Unlock Deck
Unlock for access to all 15 flashcards in this deck.
Unlock Deck
k this deck
11
You find a complex composed of a protein moiety and an RNA moiety that is capable of cleaving an RNA substrate. You want to know which moiety is responsible for catalysis. You perform an in vitro cleavage assay in the proper buffer conditions containing a low concentration of Mg 2+. The table below summarizes the results from performing seven separate reactions. The + symbol indicates the included reaction components. Sper-midine is a positively charged peptide not specific to this reaction.
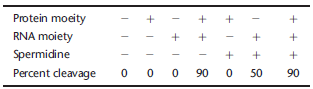
A. Does the protein or RNA moiety act as the catalyst in this reaction? Explain which reactions helped you make your conclusion.
B. Propose the function of spermidine in the last two reactions.

A. Does the protein or RNA moiety act as the catalyst in this reaction? Explain which reactions helped you make your conclusion.
B. Propose the function of spermidine in the last two reactions.

Unlock Deck
Unlock for access to all 15 flashcards in this deck.
Unlock Deck
k this deck
12
The genome of the bacteriophage Q? consists of about 4000 nucleotides of single-stranded RNA. Inside the E. coli host, replication of this genome requires a RNA-dependent RNA polymerase made up of phage and bacterial proteins. Interestingly, the replicase binds to a region in the center of the RNA genome, yet must start copying the 3? end of the RNA template to produce a new strand of RNA in the 5? to 3? direction. Researchers hypothesized that the presence of a predicted pseudoknot in the Q? genome allowed the replicase to gain access to the 3? end of the RNA. To test their hypothesis, they measured the replication efficiency in vitro using wild-type replicase and different versions of the Q? RNA containing mutations in a region key to the predicted pseudoknot formation. They specifically focused on an eight-nucleotide region from the center of the RNA that was complementary to the 3?-terminal hairpin. The wild-type and mutant sequences and replication data are given below.


In vitro replication efficiency (relative to wild type)
Wild type: 100%
Mutant A: 1.6%
Mutant B: 0.6%
Mutant C: 42%
A. Predict why the replication efficiency so low in Mutant A.
B. Predict why the replication efficiency is restored to almost half of wild-type levels in Mutant C?
C. Do you think the results support or refute the hypothesis that the presence of a pseudoknot affects replication?
Data adapted from Klovins and van Duin (1999. J. Mol. Biol. 294 ? 875-884.)


In vitro replication efficiency (relative to wild type)
Wild type: 100%
Mutant A: 1.6%
Mutant B: 0.6%
Mutant C: 42%
A. Predict why the replication efficiency so low in Mutant A.
B. Predict why the replication efficiency is restored to almost half of wild-type levels in Mutant C?
C. Do you think the results support or refute the hypothesis that the presence of a pseudoknot affects replication?
Data adapted from Klovins and van Duin (1999. J. Mol. Biol. 294 ? 875-884.)

Unlock Deck
Unlock for access to all 15 flashcards in this deck.
Unlock Deck
k this deck
13
Draw the structure of deoxyguanosine. Circle and label the appropriate hydrogen bond donors (D) or acceptors (A) that participate in hydrogen bonding when a base pair with deox-ycytidine forms in DNA.
B. Draw the structure of guanosine. Circle and label the appropriate hydrogen bond donors (D) or acceptors (A) that participate in hydrogen bonding when a base pair with uridine forms in RNA.
B. Draw the structure of guanosine. Circle and label the appropriate hydrogen bond donors (D) or acceptors (A) that participate in hydrogen bonding when a base pair with uridine forms in RNA.

Unlock Deck
Unlock for access to all 15 flashcards in this deck.
Unlock Deck
k this deck
14
Researchers discovered a new virus and characterized its genome by determining the base composition and the percentage of each base. You want to categorize the virus by its genetic material. Remember that viruses may contain single-stranded DNA or RNA or double-stranded DNA or RNA as the genetic material. Given just the sequence information, propose a way to distinguish if the genetic material is RNA or DNA. Propose a way to distinguish if the genetic information is single-stranded or double-stranded.

Unlock Deck
Unlock for access to all 15 flashcards in this deck.
Unlock Deck
k this deck
15
Explain why the helical structure of DNA differs from the helical structure of double-stranded RNA and how that difference in structure affects the ability of proteins to interact with helical RNA.

Unlock Deck
Unlock for access to all 15 flashcards in this deck.
Unlock Deck
k this deck



