Deck 3: Biology
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/319
Play
Full screen (f)
Deck 3: Biology
1
Passage
The lumen of the human gut is lined by a monolayer of epithelial cells that acts as a selectively permeable barrier, preventing the passage of harmful intraluminal foreign antigens, flora, and toxins into the circulation while allowing digestion and absorption of essential dietary nutrients along with the transfer of electrolytes and water.Proteins in the tight junctions of intestinal epithelial cells maintain barrier integrity, but barrier dysfunction occurs when these cells are damaged in the setting of infection, burns, shock, or hypoxia (low oxygen levels). The transcription factor HIF-1, a heterodimer composed of the macromolecules HIF-1α and HIF-1β, regulates the adaptive cellular response to hypoxia and the consequent expression of tight junction proteins.Researchers assessed the concentration of HIF-1 heterodimer components in human intestinal Caco-2 cells subjected to hypoxia/reoxygenation (H/R) in vitro. Caco-2 cells, a colon-derived cell line, were cultured under specific conditions to mimic the functional and morphological phenotype of wild-type enterocytes lining the small intestine. These cells were prepared and grown as a monolayer on a collagen-coated membrane.Next, the monolayer was cultured in hypoxic conditions and then exposed to atmospheric oxygen levels (normoxia) for 30, 60, and 120 minutes. Protein levels were quantified using direct enzyme-linked immunosorbent assay (ELISA), in which an antibody linked to a reporter enzyme was utilized to bind and detect expression of the target molecule (analyte) in a sample. When the colorless substrate of the reporter enzyme was added, the enzyme generated a visible colored product that could be quantified based on color intensity.
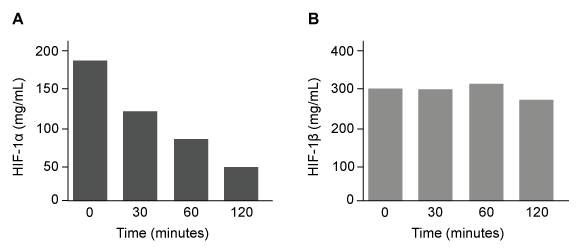 Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Adapted from Lei Q, Qiang F, Chao D, et al. Int J Mol Med. 2014;34(6):1629-39.
When Caco-2 cells are cultured in hypoxic conditions, HIF-1 is most likely located in the:
A)tight junctions.
B)cytoplasm.
C)nucleus.
D)lysosomes.
The lumen of the human gut is lined by a monolayer of epithelial cells that acts as a selectively permeable barrier, preventing the passage of harmful intraluminal foreign antigens, flora, and toxins into the circulation while allowing digestion and absorption of essential dietary nutrients along with the transfer of electrolytes and water.Proteins in the tight junctions of intestinal epithelial cells maintain barrier integrity, but barrier dysfunction occurs when these cells are damaged in the setting of infection, burns, shock, or hypoxia (low oxygen levels). The transcription factor HIF-1, a heterodimer composed of the macromolecules HIF-1α and HIF-1β, regulates the adaptive cellular response to hypoxia and the consequent expression of tight junction proteins.Researchers assessed the concentration of HIF-1 heterodimer components in human intestinal Caco-2 cells subjected to hypoxia/reoxygenation (H/R) in vitro. Caco-2 cells, a colon-derived cell line, were cultured under specific conditions to mimic the functional and morphological phenotype of wild-type enterocytes lining the small intestine. These cells were prepared and grown as a monolayer on a collagen-coated membrane.Next, the monolayer was cultured in hypoxic conditions and then exposed to atmospheric oxygen levels (normoxia) for 30, 60, and 120 minutes. Protein levels were quantified using direct enzyme-linked immunosorbent assay (ELISA), in which an antibody linked to a reporter enzyme was utilized to bind and detect expression of the target molecule (analyte) in a sample. When the colorless substrate of the reporter enzyme was added, the enzyme generated a visible colored product that could be quantified based on color intensity.
 Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.Adapted from Lei Q, Qiang F, Chao D, et al. Int J Mol Med. 2014;34(6):1629-39.
When Caco-2 cells are cultured in hypoxic conditions, HIF-1 is most likely located in the:
A)tight junctions.
B)cytoplasm.
C)nucleus.
D)lysosomes.
nucleus.
2
Passage
Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 105 SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.Table 1 Mean anti-SRC titers with standard error
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. Experimental results suggest that the mutated Tlr4 allele exhibits which quality?</strong> A)Codominance B)Recessivity C)Reduced penetrance D)Variable expressivity](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_2c1d_a3bf_85095183ef29_MD0008_00.jpg) Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. Experimental results suggest that the mutated Tlr4 allele exhibits which quality?</strong> A)Codominance B)Recessivity C)Reduced penetrance D)Variable expressivity](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_532e_a3bf_2bc230da9462_MD0008_00.jpg) Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
Experimental results suggest that the mutated Tlr4 allele exhibits which quality?
A)Codominance
B)Recessivity
C)Reduced penetrance
D)Variable expressivity
Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 105 SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.Table 1 Mean anti-SRC titers with standard error
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. Experimental results suggest that the mutated Tlr4 allele exhibits which quality?</strong> A)Codominance B)Recessivity C)Reduced penetrance D)Variable expressivity](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_2c1d_a3bf_85095183ef29_MD0008_00.jpg) Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. Experimental results suggest that the mutated Tlr4 allele exhibits which quality?</strong> A)Codominance B)Recessivity C)Reduced penetrance D)Variable expressivity](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_532e_a3bf_2bc230da9462_MD0008_00.jpg) Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.Experimental results suggest that the mutated Tlr4 allele exhibits which quality?
A)Codominance
B)Recessivity
C)Reduced penetrance
D)Variable expressivity
Codominance
3
Passage
At the blood-brain barrier, the cerebral spinal fluid (CSF) is separated from lymphatic circulation by epithelial cells bound by tight junctions. Within the central nervous system (CNS), CSF surrounds and protects the brain and spinal cord and is believed to function in waste clearance for the CNS analogous to the role of the lymphatic system within the body.This waste clearance system has been dubbed the "glymphatic system." Although the mechanism of clearance is largely unknown, specialized glial cells known as astrocytes appear to modify the interstitial volume between neurons. This increase in interstitial volume allows for greater CSF flow, which increases the efficiency of neurotoxic clearance. Waste products removed from the brain include metabolic products such as ammonia and harmful compounds such as amyloid proteins.Recent advances in imaging technology suggest that interstitial clearance may be modified during sleep. To test this hypothesis, researchers indirectly studied changes in interstitial volume by measuring the percentage of CSF coverage in animal models during the first hour of sleep and again during the first hour of wakefulness (Figure 1).
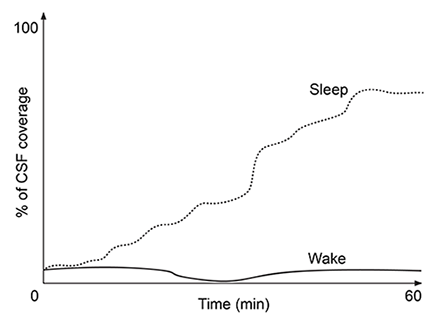 Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
A patient ingests the following pharmaceutical compound prior to sleep.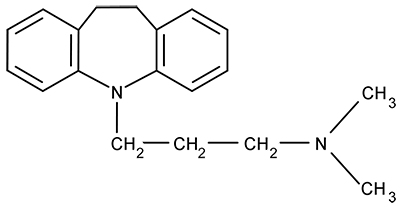 During the hours following ingestion, the drug concentration in the brain will:
During the hours following ingestion, the drug concentration in the brain will:
A)increase less rapidly as interstitial volume increases.
B)increase more rapidly as interstitial volume increases.
C)increase independently of interstitial volume changes.
D)remain zero regardless of interstitial volume.
At the blood-brain barrier, the cerebral spinal fluid (CSF) is separated from lymphatic circulation by epithelial cells bound by tight junctions. Within the central nervous system (CNS), CSF surrounds and protects the brain and spinal cord and is believed to function in waste clearance for the CNS analogous to the role of the lymphatic system within the body.This waste clearance system has been dubbed the "glymphatic system." Although the mechanism of clearance is largely unknown, specialized glial cells known as astrocytes appear to modify the interstitial volume between neurons. This increase in interstitial volume allows for greater CSF flow, which increases the efficiency of neurotoxic clearance. Waste products removed from the brain include metabolic products such as ammonia and harmful compounds such as amyloid proteins.Recent advances in imaging technology suggest that interstitial clearance may be modified during sleep. To test this hypothesis, researchers indirectly studied changes in interstitial volume by measuring the percentage of CSF coverage in animal models during the first hour of sleep and again during the first hour of wakefulness (Figure 1).
 Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulnessA patient ingests the following pharmaceutical compound prior to sleep.
 During the hours following ingestion, the drug concentration in the brain will:
During the hours following ingestion, the drug concentration in the brain will:A)increase less rapidly as interstitial volume increases.
B)increase more rapidly as interstitial volume increases.
C)increase independently of interstitial volume changes.
D)remain zero regardless of interstitial volume.
increase less rapidly as interstitial volume increases.
4
Passage
Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 105 SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.Table 1 Mean anti-SRC titers with standard error
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. If researchers repeated the analyses using F2 generation mice, spleen cell cultures exposed to SRC plus 1.0 μg Ph-LPS would be expected to have anti-SRC titers of:16001200800</strong> A)II only B)I and II only C)I and III only D)I, II, and III](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_2c1d_a3bf_85095183ef29_MD0008_00.jpg) Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. If researchers repeated the analyses using F2 generation mice, spleen cell cultures exposed to SRC plus 1.0 μg Ph-LPS would be expected to have anti-SRC titers of:16001200800</strong> A)II only B)I and II only C)I and III only D)I, II, and III](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_532e_a3bf_2bc230da9462_MD0008_00.jpg) Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
If researchers repeated the analyses using F2 generation mice, spleen cell cultures exposed to SRC plus 1.0 μg Ph-LPS would be expected to have anti-SRC titers of:16001200800
A)II only
B)I and II only
C)I and III only
D)I, II, and III
Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 105 SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.Table 1 Mean anti-SRC titers with standard error
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. If researchers repeated the analyses using F2 generation mice, spleen cell cultures exposed to SRC plus 1.0 μg Ph-LPS would be expected to have anti-SRC titers of:16001200800</strong> A)II only B)I and II only C)I and III only D)I, II, and III](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_2c1d_a3bf_85095183ef29_MD0008_00.jpg) Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. If researchers repeated the analyses using F2 generation mice, spleen cell cultures exposed to SRC plus 1.0 μg Ph-LPS would be expected to have anti-SRC titers of:16001200800</strong> A)II only B)I and II only C)I and III only D)I, II, and III](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_532e_a3bf_2bc230da9462_MD0008_00.jpg) Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.If researchers repeated the analyses using F2 generation mice, spleen cell cultures exposed to SRC plus 1.0 μg Ph-LPS would be expected to have anti-SRC titers of:16001200800
A)II only
B)I and II only
C)I and III only
D)I, II, and III

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
5
Passage
At the blood-brain barrier, the cerebral spinal fluid (CSF) is separated from lymphatic circulation by epithelial cells bound by tight junctions. Within the central nervous system (CNS), CSF surrounds and protects the brain and spinal cord and is believed to function in waste clearance for the CNS analogous to the role of the lymphatic system within the body.This waste clearance system has been dubbed the "glymphatic system." Although the mechanism of clearance is largely unknown, specialized glial cells known as astrocytes appear to modify the interstitial volume between neurons. This increase in interstitial volume allows for greater CSF flow, which increases the efficiency of neurotoxic clearance. Waste products removed from the brain include metabolic products such as ammonia and harmful compounds such as amyloid proteins.Recent advances in imaging technology suggest that interstitial clearance may be modified during sleep. To test this hypothesis, researchers indirectly studied changes in interstitial volume by measuring the percentage of CSF coverage in animal models during the first hour of sleep and again during the first hour of wakefulness (Figure 1).
 Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
A second study is performed to measure CSF coverage against brain wave frequency, measured by electroencephalography. Which of the following findings would best support the data provided in the passage?
A)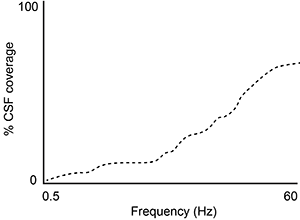
B)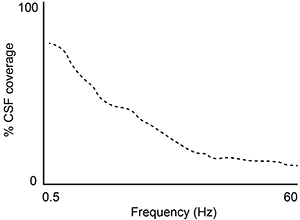
C)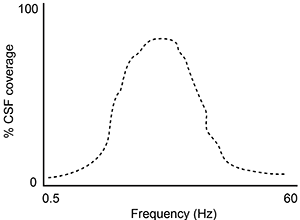
D)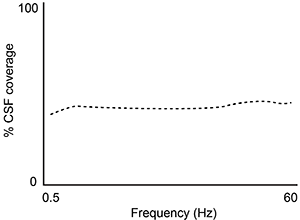
At the blood-brain barrier, the cerebral spinal fluid (CSF) is separated from lymphatic circulation by epithelial cells bound by tight junctions. Within the central nervous system (CNS), CSF surrounds and protects the brain and spinal cord and is believed to function in waste clearance for the CNS analogous to the role of the lymphatic system within the body.This waste clearance system has been dubbed the "glymphatic system." Although the mechanism of clearance is largely unknown, specialized glial cells known as astrocytes appear to modify the interstitial volume between neurons. This increase in interstitial volume allows for greater CSF flow, which increases the efficiency of neurotoxic clearance. Waste products removed from the brain include metabolic products such as ammonia and harmful compounds such as amyloid proteins.Recent advances in imaging technology suggest that interstitial clearance may be modified during sleep. To test this hypothesis, researchers indirectly studied changes in interstitial volume by measuring the percentage of CSF coverage in animal models during the first hour of sleep and again during the first hour of wakefulness (Figure 1).
 Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulnessA second study is performed to measure CSF coverage against brain wave frequency, measured by electroencephalography. Which of the following findings would best support the data provided in the passage?
A)

B)

C)

D)


Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
6
Passage
The lumen of the human gut is lined by a monolayer of epithelial cells that acts as a selectively permeable barrier, preventing the passage of harmful intraluminal foreign antigens, flora, and toxins into the circulation while allowing digestion and absorption of essential dietary nutrients along with the transfer of electrolytes and water.Proteins in the tight junctions of intestinal epithelial cells maintain barrier integrity, but barrier dysfunction occurs when these cells are damaged in the setting of infection, burns, shock, or hypoxia (low oxygen levels). The transcription factor HIF-1, a heterodimer composed of the macromolecules HIF-1α and HIF-1β, regulates the adaptive cellular response to hypoxia and the consequent expression of tight junction proteins.Researchers assessed the concentration of HIF-1 heterodimer components in human intestinal Caco-2 cells subjected to hypoxia/reoxygenation (H/R) in vitro. Caco-2 cells, a colon-derived cell line, were cultured under specific conditions to mimic the functional and morphological phenotype of wild-type enterocytes lining the small intestine. These cells were prepared and grown as a monolayer on a collagen-coated membrane.Next, the monolayer was cultured in hypoxic conditions and then exposed to atmospheric oxygen levels (normoxia) for 30, 60, and 120 minutes. Protein levels were quantified using direct enzyme-linked immunosorbent assay (ELISA), in which an antibody linked to a reporter enzyme was utilized to bind and detect expression of the target molecule (analyte) in a sample. When the colorless substrate of the reporter enzyme was added, the enzyme generated a visible colored product that could be quantified based on color intensity.
 Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Adapted from Lei Q, Qiang F, Chao D, et al. Int J Mol Med. 2014;34(6):1629-39.
Based on the passage, Caco-2 cells originate from a segment of the human gut that mainly functions to:
A)absorb nutrients.
B)absorb water.
C)produce proteolytic enzymes.
D)digest nutrients.
The lumen of the human gut is lined by a monolayer of epithelial cells that acts as a selectively permeable barrier, preventing the passage of harmful intraluminal foreign antigens, flora, and toxins into the circulation while allowing digestion and absorption of essential dietary nutrients along with the transfer of electrolytes and water.Proteins in the tight junctions of intestinal epithelial cells maintain barrier integrity, but barrier dysfunction occurs when these cells are damaged in the setting of infection, burns, shock, or hypoxia (low oxygen levels). The transcription factor HIF-1, a heterodimer composed of the macromolecules HIF-1α and HIF-1β, regulates the adaptive cellular response to hypoxia and the consequent expression of tight junction proteins.Researchers assessed the concentration of HIF-1 heterodimer components in human intestinal Caco-2 cells subjected to hypoxia/reoxygenation (H/R) in vitro. Caco-2 cells, a colon-derived cell line, were cultured under specific conditions to mimic the functional and morphological phenotype of wild-type enterocytes lining the small intestine. These cells were prepared and grown as a monolayer on a collagen-coated membrane.Next, the monolayer was cultured in hypoxic conditions and then exposed to atmospheric oxygen levels (normoxia) for 30, 60, and 120 minutes. Protein levels were quantified using direct enzyme-linked immunosorbent assay (ELISA), in which an antibody linked to a reporter enzyme was utilized to bind and detect expression of the target molecule (analyte) in a sample. When the colorless substrate of the reporter enzyme was added, the enzyme generated a visible colored product that could be quantified based on color intensity.
 Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.Adapted from Lei Q, Qiang F, Chao D, et al. Int J Mol Med. 2014;34(6):1629-39.
Based on the passage, Caco-2 cells originate from a segment of the human gut that mainly functions to:
A)absorb nutrients.
B)absorb water.
C)produce proteolytic enzymes.
D)digest nutrients.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
7
Passage
Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure.
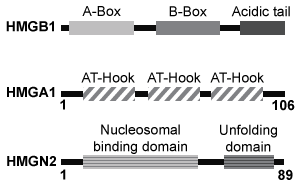 Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
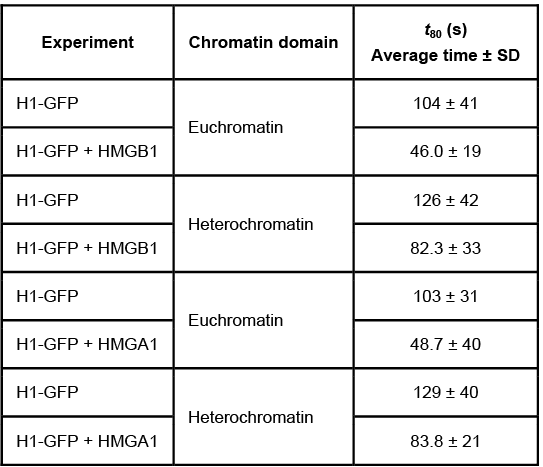 Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
The experimental design described in the passage allowed researchers to do which of the following?Assess whether structural chromatin organization influences H1 mobilityAssess H1 and GFP competition at different levels of DNA compactionEvaluate if HMG proteins compete with H1 for chromatin-binding sites
A)I and III only
B)I and II only
C)II and III only
D)I, II, and III
Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure.
 Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.The experimental design described in the passage allowed researchers to do which of the following?Assess whether structural chromatin organization influences H1 mobilityAssess H1 and GFP competition at different levels of DNA compactionEvaluate if HMG proteins compete with H1 for chromatin-binding sites
A)I and III only
B)I and II only
C)II and III only
D)I, II, and III

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
8
Passage
Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 105 SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.Table 1 Mean anti-SRC titers with standard error
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. The experiments allowed researchers to determine the effect of genotype variation on all of the following EXCEPT:</strong> A)in vitro versus in vivo studies. B)innate versus adaptive immune responses. C)responses modulated by LPS exposure. D)qualitative and quantitative phenotypic traits.](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_2c1d_a3bf_85095183ef29_MD0008_00.jpg) Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. The experiments allowed researchers to determine the effect of genotype variation on all of the following EXCEPT:</strong> A)in vitro versus in vivo studies. B)innate versus adaptive immune responses. C)responses modulated by LPS exposure. D)qualitative and quantitative phenotypic traits.](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_532e_a3bf_2bc230da9462_MD0008_00.jpg) Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
The experiments allowed researchers to determine the effect of genotype variation on all of the following EXCEPT:
A)in vitro versus in vivo studies.
B)innate versus adaptive immune responses.
C)responses modulated by LPS exposure.
D)qualitative and quantitative phenotypic traits.
Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 105 SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.Table 1 Mean anti-SRC titers with standard error
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. The experiments allowed researchers to determine the effect of genotype variation on all of the following EXCEPT:</strong> A)in vitro versus in vivo studies. B)innate versus adaptive immune responses. C)responses modulated by LPS exposure. D)qualitative and quantitative phenotypic traits.](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_2c1d_a3bf_85095183ef29_MD0008_00.jpg) Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. The experiments allowed researchers to determine the effect of genotype variation on all of the following EXCEPT:</strong> A)in vitro versus in vivo studies. B)innate versus adaptive immune responses. C)responses modulated by LPS exposure. D)qualitative and quantitative phenotypic traits.](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_532e_a3bf_2bc230da9462_MD0008_00.jpg) Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.The experiments allowed researchers to determine the effect of genotype variation on all of the following EXCEPT:
A)in vitro versus in vivo studies.
B)innate versus adaptive immune responses.
C)responses modulated by LPS exposure.
D)qualitative and quantitative phenotypic traits.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
9
Passage
Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 105 SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.Table 1 Mean anti-SRC titers with standard error
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. In an isolated population of 10,000 mice, 1,600 are homozygous for a Tlr4 defect. Assuming stable allele and genotype frequencies, how many mice are heterozygous for a Tlr4 defect?</strong> A)2,400 B)3,600 C)4,800 D)5,200](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_2c1d_a3bf_85095183ef29_MD0008_00.jpg) Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. In an isolated population of 10,000 mice, 1,600 are homozygous for a Tlr4 defect. Assuming stable allele and genotype frequencies, how many mice are heterozygous for a Tlr4 defect?</strong> A)2,400 B)3,600 C)4,800 D)5,200](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_532e_a3bf_2bc230da9462_MD0008_00.jpg) Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
In an isolated population of 10,000 mice, 1,600 are homozygous for a Tlr4 defect. Assuming stable allele and genotype frequencies, how many mice are heterozygous for a Tlr4 defect?
A)2,400
B)3,600
C)4,800
D)5,200
Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 105 SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.Table 1 Mean anti-SRC titers with standard error
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. In an isolated population of 10,000 mice, 1,600 are homozygous for a Tlr4 defect. Assuming stable allele and genotype frequencies, how many mice are heterozygous for a Tlr4 defect?</strong> A)2,400 B)3,600 C)4,800 D)5,200](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_2c1d_a3bf_85095183ef29_MD0008_00.jpg) Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. In an isolated population of 10,000 mice, 1,600 are homozygous for a Tlr4 defect. Assuming stable allele and genotype frequencies, how many mice are heterozygous for a Tlr4 defect?</strong> A)2,400 B)3,600 C)4,800 D)5,200](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_532e_a3bf_2bc230da9462_MD0008_00.jpg) Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.In an isolated population of 10,000 mice, 1,600 are homozygous for a Tlr4 defect. Assuming stable allele and genotype frequencies, how many mice are heterozygous for a Tlr4 defect?
A)2,400
B)3,600
C)4,800
D)5,200

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
10
Passage
The lumen of the human gut is lined by a monolayer of epithelial cells that acts as a selectively permeable barrier, preventing the passage of harmful intraluminal foreign antigens, flora, and toxins into the circulation while allowing digestion and absorption of essential dietary nutrients along with the transfer of electrolytes and water.Proteins in the tight junctions of intestinal epithelial cells maintain barrier integrity, but barrier dysfunction occurs when these cells are damaged in the setting of infection, burns, shock, or hypoxia (low oxygen levels). The transcription factor HIF-1, a heterodimer composed of the macromolecules HIF-1α and HIF-1β, regulates the adaptive cellular response to hypoxia and the consequent expression of tight junction proteins.Researchers assessed the concentration of HIF-1 heterodimer components in human intestinal Caco-2 cells subjected to hypoxia/reoxygenation (H/R) in vitro. Caco-2 cells, a colon-derived cell line, were cultured under specific conditions to mimic the functional and morphological phenotype of wild-type enterocytes lining the small intestine. These cells were prepared and grown as a monolayer on a collagen-coated membrane.Next, the monolayer was cultured in hypoxic conditions and then exposed to atmospheric oxygen levels (normoxia) for 30, 60, and 120 minutes. Protein levels were quantified using direct enzyme-linked immunosorbent assay (ELISA), in which an antibody linked to a reporter enzyme was utilized to bind and detect expression of the target molecule (analyte) in a sample. When the colorless substrate of the reporter enzyme was added, the enzyme generated a visible colored product that could be quantified based on color intensity.
 Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Adapted from Lei Q, Qiang F, Chao D, et al. Int J Mol Med. 2014;34(6):1629-39.
Developmental protocols for Caco-2 monolayers involve the use of cells with high proliferation potential that can differentiate in a synchronized manner to form homogenous monolayers. This proliferation of Caco-2 cells is primarily achieved through:
A)meiosis.
B)fission.
C)completion of the G2 phase of the cell cycle.
D)completion of the M phase of the cell cycle.
The lumen of the human gut is lined by a monolayer of epithelial cells that acts as a selectively permeable barrier, preventing the passage of harmful intraluminal foreign antigens, flora, and toxins into the circulation while allowing digestion and absorption of essential dietary nutrients along with the transfer of electrolytes and water.Proteins in the tight junctions of intestinal epithelial cells maintain barrier integrity, but barrier dysfunction occurs when these cells are damaged in the setting of infection, burns, shock, or hypoxia (low oxygen levels). The transcription factor HIF-1, a heterodimer composed of the macromolecules HIF-1α and HIF-1β, regulates the adaptive cellular response to hypoxia and the consequent expression of tight junction proteins.Researchers assessed the concentration of HIF-1 heterodimer components in human intestinal Caco-2 cells subjected to hypoxia/reoxygenation (H/R) in vitro. Caco-2 cells, a colon-derived cell line, were cultured under specific conditions to mimic the functional and morphological phenotype of wild-type enterocytes lining the small intestine. These cells were prepared and grown as a monolayer on a collagen-coated membrane.Next, the monolayer was cultured in hypoxic conditions and then exposed to atmospheric oxygen levels (normoxia) for 30, 60, and 120 minutes. Protein levels were quantified using direct enzyme-linked immunosorbent assay (ELISA), in which an antibody linked to a reporter enzyme was utilized to bind and detect expression of the target molecule (analyte) in a sample. When the colorless substrate of the reporter enzyme was added, the enzyme generated a visible colored product that could be quantified based on color intensity.
 Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.Adapted from Lei Q, Qiang F, Chao D, et al. Int J Mol Med. 2014;34(6):1629-39.
Developmental protocols for Caco-2 monolayers involve the use of cells with high proliferation potential that can differentiate in a synchronized manner to form homogenous monolayers. This proliferation of Caco-2 cells is primarily achieved through:
A)meiosis.
B)fission.
C)completion of the G2 phase of the cell cycle.
D)completion of the M phase of the cell cycle.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
11
Passage
At the blood-brain barrier, the cerebral spinal fluid (CSF) is separated from lymphatic circulation by epithelial cells bound by tight junctions. Within the central nervous system (CNS), CSF surrounds and protects the brain and spinal cord and is believed to function in waste clearance for the CNS analogous to the role of the lymphatic system within the body.This waste clearance system has been dubbed the "glymphatic system." Although the mechanism of clearance is largely unknown, specialized glial cells known as astrocytes appear to modify the interstitial volume between neurons. This increase in interstitial volume allows for greater CSF flow, which increases the efficiency of neurotoxic clearance. Waste products removed from the brain include metabolic products such as ammonia and harmful compounds such as amyloid proteins.Recent advances in imaging technology suggest that interstitial clearance may be modified during sleep. To test this hypothesis, researchers indirectly studied changes in interstitial volume by measuring the percentage of CSF coverage in animal models during the first hour of sleep and again during the first hour of wakefulness (Figure 1).
 Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Which of the following is true regarding the function of neuroglia in the central and peripheral nervous systems?
A)Astrocytes are the epithelial cells of the central nervous system and secrete CSF.
B)Each Schwann cell myelinates multiple axons in the peripheral nervous system.
C)Each oligodendrocyte myelinates a single axon in the peripheral nervous system.
D)Microglia are the macrophages of the central nervous system and consume waste.
At the blood-brain barrier, the cerebral spinal fluid (CSF) is separated from lymphatic circulation by epithelial cells bound by tight junctions. Within the central nervous system (CNS), CSF surrounds and protects the brain and spinal cord and is believed to function in waste clearance for the CNS analogous to the role of the lymphatic system within the body.This waste clearance system has been dubbed the "glymphatic system." Although the mechanism of clearance is largely unknown, specialized glial cells known as astrocytes appear to modify the interstitial volume between neurons. This increase in interstitial volume allows for greater CSF flow, which increases the efficiency of neurotoxic clearance. Waste products removed from the brain include metabolic products such as ammonia and harmful compounds such as amyloid proteins.Recent advances in imaging technology suggest that interstitial clearance may be modified during sleep. To test this hypothesis, researchers indirectly studied changes in interstitial volume by measuring the percentage of CSF coverage in animal models during the first hour of sleep and again during the first hour of wakefulness (Figure 1).
 Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulnessWhich of the following is true regarding the function of neuroglia in the central and peripheral nervous systems?
A)Astrocytes are the epithelial cells of the central nervous system and secrete CSF.
B)Each Schwann cell myelinates multiple axons in the peripheral nervous system.
C)Each oligodendrocyte myelinates a single axon in the peripheral nervous system.
D)Microglia are the macrophages of the central nervous system and consume waste.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
12
Passage
Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure.
 Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
 Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
A recent study of a model organism showed that HMG-5, another member of the HMG family, mainly localized to the end of chromosomes. The site of HMG-5 localization would correspond to regions in humans that:
A)contain multiple copies of different DNA sequences.
B)contain heterochromatin similar to that of centromeres.
C)are subsequently lengthened with each round of cell division.
D)are actively replicated by the enzyme DNA polymerase.
Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure.
 Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.A recent study of a model organism showed that HMG-5, another member of the HMG family, mainly localized to the end of chromosomes. The site of HMG-5 localization would correspond to regions in humans that:
A)contain multiple copies of different DNA sequences.
B)contain heterochromatin similar to that of centromeres.
C)are subsequently lengthened with each round of cell division.
D)are actively replicated by the enzyme DNA polymerase.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
13
Passage
At the blood-brain barrier, the cerebral spinal fluid (CSF) is separated from lymphatic circulation by epithelial cells bound by tight junctions. Within the central nervous system (CNS), CSF surrounds and protects the brain and spinal cord and is believed to function in waste clearance for the CNS analogous to the role of the lymphatic system within the body.This waste clearance system has been dubbed the "glymphatic system." Although the mechanism of clearance is largely unknown, specialized glial cells known as astrocytes appear to modify the interstitial volume between neurons. This increase in interstitial volume allows for greater CSF flow, which increases the efficiency of neurotoxic clearance. Waste products removed from the brain include metabolic products such as ammonia and harmful compounds such as amyloid proteins.Recent advances in imaging technology suggest that interstitial clearance may be modified during sleep. To test this hypothesis, researchers indirectly studied changes in interstitial volume by measuring the percentage of CSF coverage in animal models during the first hour of sleep and again during the first hour of wakefulness (Figure 1).
 Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Which of the following sequences accurately describes the pathway of communication between neurons?
A)Axon, soma, dendrite, synapse
B)Axon, synapse, dendrite, soma
C)Dendrite, axon, soma, synapse
D)Synapse, soma, axon, dendrite
At the blood-brain barrier, the cerebral spinal fluid (CSF) is separated from lymphatic circulation by epithelial cells bound by tight junctions. Within the central nervous system (CNS), CSF surrounds and protects the brain and spinal cord and is believed to function in waste clearance for the CNS analogous to the role of the lymphatic system within the body.This waste clearance system has been dubbed the "glymphatic system." Although the mechanism of clearance is largely unknown, specialized glial cells known as astrocytes appear to modify the interstitial volume between neurons. This increase in interstitial volume allows for greater CSF flow, which increases the efficiency of neurotoxic clearance. Waste products removed from the brain include metabolic products such as ammonia and harmful compounds such as amyloid proteins.Recent advances in imaging technology suggest that interstitial clearance may be modified during sleep. To test this hypothesis, researchers indirectly studied changes in interstitial volume by measuring the percentage of CSF coverage in animal models during the first hour of sleep and again during the first hour of wakefulness (Figure 1).
 Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulnessWhich of the following sequences accurately describes the pathway of communication between neurons?
A)Axon, soma, dendrite, synapse
B)Axon, synapse, dendrite, soma
C)Dendrite, axon, soma, synapse
D)Synapse, soma, axon, dendrite

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
14
Passage
Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure.
 Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
 Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
Based on Figure 1, HMGN2 is most likely to interact with a histone complex that is rich in:
A)aspartate and glutamate.
B)aspartate and arginine.
C)lysine and glutamate.
D)lysine and arginine.
Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure.
 Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.Based on Figure 1, HMGN2 is most likely to interact with a histone complex that is rich in:
A)aspartate and glutamate.
B)aspartate and arginine.
C)lysine and glutamate.
D)lysine and arginine.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
15
Passage
The lumen of the human gut is lined by a monolayer of epithelial cells that acts as a selectively permeable barrier, preventing the passage of harmful intraluminal foreign antigens, flora, and toxins into the circulation while allowing digestion and absorption of essential dietary nutrients along with the transfer of electrolytes and water.Proteins in the tight junctions of intestinal epithelial cells maintain barrier integrity, but barrier dysfunction occurs when these cells are damaged in the setting of infection, burns, shock, or hypoxia (low oxygen levels). The transcription factor HIF-1, a heterodimer composed of the macromolecules HIF-1α and HIF-1β, regulates the adaptive cellular response to hypoxia and the consequent expression of tight junction proteins.Researchers assessed the concentration of HIF-1 heterodimer components in human intestinal Caco-2 cells subjected to hypoxia/reoxygenation (H/R) in vitro. Caco-2 cells, a colon-derived cell line, were cultured under specific conditions to mimic the functional and morphological phenotype of wild-type enterocytes lining the small intestine. These cells were prepared and grown as a monolayer on a collagen-coated membrane.Next, the monolayer was cultured in hypoxic conditions and then exposed to atmospheric oxygen levels (normoxia) for 30, 60, and 120 minutes. Protein levels were quantified using direct enzyme-linked immunosorbent assay (ELISA), in which an antibody linked to a reporter enzyme was utilized to bind and detect expression of the target molecule (analyte) in a sample. When the colorless substrate of the reporter enzyme was added, the enzyme generated a visible colored product that could be quantified based on color intensity.
 Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Adapted from Lei Q, Qiang F, Chao D, et al. Int J Mol Med. 2014;34(6):1629-39.
Which graphic depicts the most likely effect of the HIF-1α inhibitor on emodin? (Note: control = cells cultured under normoxic conditions.)
A)
B)
C)
D)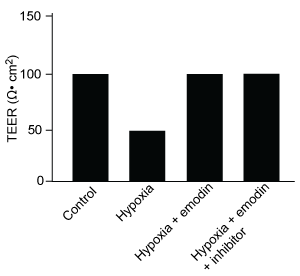
The lumen of the human gut is lined by a monolayer of epithelial cells that acts as a selectively permeable barrier, preventing the passage of harmful intraluminal foreign antigens, flora, and toxins into the circulation while allowing digestion and absorption of essential dietary nutrients along with the transfer of electrolytes and water.Proteins in the tight junctions of intestinal epithelial cells maintain barrier integrity, but barrier dysfunction occurs when these cells are damaged in the setting of infection, burns, shock, or hypoxia (low oxygen levels). The transcription factor HIF-1, a heterodimer composed of the macromolecules HIF-1α and HIF-1β, regulates the adaptive cellular response to hypoxia and the consequent expression of tight junction proteins.Researchers assessed the concentration of HIF-1 heterodimer components in human intestinal Caco-2 cells subjected to hypoxia/reoxygenation (H/R) in vitro. Caco-2 cells, a colon-derived cell line, were cultured under specific conditions to mimic the functional and morphological phenotype of wild-type enterocytes lining the small intestine. These cells were prepared and grown as a monolayer on a collagen-coated membrane.Next, the monolayer was cultured in hypoxic conditions and then exposed to atmospheric oxygen levels (normoxia) for 30, 60, and 120 minutes. Protein levels were quantified using direct enzyme-linked immunosorbent assay (ELISA), in which an antibody linked to a reporter enzyme was utilized to bind and detect expression of the target molecule (analyte) in a sample. When the colorless substrate of the reporter enzyme was added, the enzyme generated a visible colored product that could be quantified based on color intensity.
 Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.Adapted from Lei Q, Qiang F, Chao D, et al. Int J Mol Med. 2014;34(6):1629-39.
Which graphic depicts the most likely effect of the HIF-1α inhibitor on emodin? (Note: control = cells cultured under normoxic conditions.)
A)

B)

C)

D)


Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
16
Passage
At the blood-brain barrier, the cerebral spinal fluid (CSF) is separated from lymphatic circulation by epithelial cells bound by tight junctions. Within the central nervous system (CNS), CSF surrounds and protects the brain and spinal cord and is believed to function in waste clearance for the CNS analogous to the role of the lymphatic system within the body.This waste clearance system has been dubbed the "glymphatic system." Although the mechanism of clearance is largely unknown, specialized glial cells known as astrocytes appear to modify the interstitial volume between neurons. This increase in interstitial volume allows for greater CSF flow, which increases the efficiency of neurotoxic clearance. Waste products removed from the brain include metabolic products such as ammonia and harmful compounds such as amyloid proteins.Recent advances in imaging technology suggest that interstitial clearance may be modified during sleep. To test this hypothesis, researchers indirectly studied changes in interstitial volume by measuring the percentage of CSF coverage in animal models during the first hour of sleep and again during the first hour of wakefulness (Figure 1).
 Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Given the role of the glymphatic system outlined in the passage, chronic sleep deprivation might contribute to the development of which of the following disease states?
A)Alzheimer's disease
B)Multiple sclerosis
C)Huntington's disease
D)Parkinson's disease
At the blood-brain barrier, the cerebral spinal fluid (CSF) is separated from lymphatic circulation by epithelial cells bound by tight junctions. Within the central nervous system (CNS), CSF surrounds and protects the brain and spinal cord and is believed to function in waste clearance for the CNS analogous to the role of the lymphatic system within the body.This waste clearance system has been dubbed the "glymphatic system." Although the mechanism of clearance is largely unknown, specialized glial cells known as astrocytes appear to modify the interstitial volume between neurons. This increase in interstitial volume allows for greater CSF flow, which increases the efficiency of neurotoxic clearance. Waste products removed from the brain include metabolic products such as ammonia and harmful compounds such as amyloid proteins.Recent advances in imaging technology suggest that interstitial clearance may be modified during sleep. To test this hypothesis, researchers indirectly studied changes in interstitial volume by measuring the percentage of CSF coverage in animal models during the first hour of sleep and again during the first hour of wakefulness (Figure 1).
 Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulnessGiven the role of the glymphatic system outlined in the passage, chronic sleep deprivation might contribute to the development of which of the following disease states?
A)Alzheimer's disease
B)Multiple sclerosis
C)Huntington's disease
D)Parkinson's disease

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
17
Passage
At the blood-brain barrier, the cerebral spinal fluid (CSF) is separated from lymphatic circulation by epithelial cells bound by tight junctions. Within the central nervous system (CNS), CSF surrounds and protects the brain and spinal cord and is believed to function in waste clearance for the CNS analogous to the role of the lymphatic system within the body.This waste clearance system has been dubbed the "glymphatic system." Although the mechanism of clearance is largely unknown, specialized glial cells known as astrocytes appear to modify the interstitial volume between neurons. This increase in interstitial volume allows for greater CSF flow, which increases the efficiency of neurotoxic clearance. Waste products removed from the brain include metabolic products such as ammonia and harmful compounds such as amyloid proteins.Recent advances in imaging technology suggest that interstitial clearance may be modified during sleep. To test this hypothesis, researchers indirectly studied changes in interstitial volume by measuring the percentage of CSF coverage in animal models during the first hour of sleep and again during the first hour of wakefulness (Figure 1).
 Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Which of the following is true regarding the organization of neuronal tracts in the spinal cord?
A)Afferent neuronal fibers carry motor commands from the brain to the body through tracts in the spinal gray matter.
B)Afferent neuronal fibers carry sensory information from the body to the brain through tracts in the spinal gray matter.
C)Efferent neuronal fibers carry motor commands from the brain to the body through tracts in the spinal white matter.
D)Efferent neuronal fibers carry sensory information from the body to the brain through tracts in the spinal white matter.
At the blood-brain barrier, the cerebral spinal fluid (CSF) is separated from lymphatic circulation by epithelial cells bound by tight junctions. Within the central nervous system (CNS), CSF surrounds and protects the brain and spinal cord and is believed to function in waste clearance for the CNS analogous to the role of the lymphatic system within the body.This waste clearance system has been dubbed the "glymphatic system." Although the mechanism of clearance is largely unknown, specialized glial cells known as astrocytes appear to modify the interstitial volume between neurons. This increase in interstitial volume allows for greater CSF flow, which increases the efficiency of neurotoxic clearance. Waste products removed from the brain include metabolic products such as ammonia and harmful compounds such as amyloid proteins.Recent advances in imaging technology suggest that interstitial clearance may be modified during sleep. To test this hypothesis, researchers indirectly studied changes in interstitial volume by measuring the percentage of CSF coverage in animal models during the first hour of sleep and again during the first hour of wakefulness (Figure 1).
 Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulness
Figure 1. Percentage of CSF coverage of interstitial space during sleep and wakefulnessWhich of the following is true regarding the organization of neuronal tracts in the spinal cord?
A)Afferent neuronal fibers carry motor commands from the brain to the body through tracts in the spinal gray matter.
B)Afferent neuronal fibers carry sensory information from the body to the brain through tracts in the spinal gray matter.
C)Efferent neuronal fibers carry motor commands from the brain to the body through tracts in the spinal white matter.
D)Efferent neuronal fibers carry sensory information from the body to the brain through tracts in the spinal white matter.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
18
Passage
Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure.
 Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
 Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
If mutant HMGN2 proteins were unable to bind their substrate and then erroneously mobilize to nucleoli, these proteins would be found in the sites of:
A)lipid synthesis.
B)ATP synthesis.
C)ribosome production.
D)ribosome attachment.
Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure.
 Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.If mutant HMGN2 proteins were unable to bind their substrate and then erroneously mobilize to nucleoli, these proteins would be found in the sites of:
A)lipid synthesis.
B)ATP synthesis.
C)ribosome production.
D)ribosome attachment.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
19
Passage
Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure.
 Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
 Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
The molecules in Figure 1 were analyzed using gel electrophoresis, and the results are shown below. Which of the following describes the correct experimental methodology?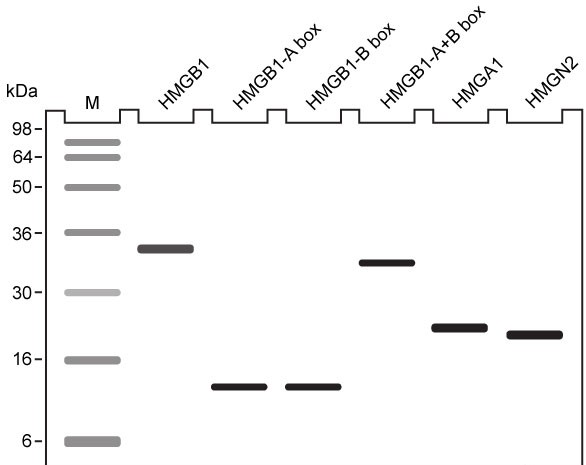
A)The intrinsic charges of the proteins were masked by a reducing agent.
B)The disulfide bonds were disrupted by the detergent sodium dodecyl sulfate.
C)The bands on the gel were stained with a dye that causes DNA to fluoresce.
D)The molecules were run through a highly crosslinked polyacrylamide gel.
Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure.
 Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.The molecules in Figure 1 were analyzed using gel electrophoresis, and the results are shown below. Which of the following describes the correct experimental methodology?

A)The intrinsic charges of the proteins were masked by a reducing agent.
B)The disulfide bonds were disrupted by the detergent sodium dodecyl sulfate.
C)The bands on the gel were stained with a dye that causes DNA to fluoresce.
D)The molecules were run through a highly crosslinked polyacrylamide gel.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
20
Passage
The lumen of the human gut is lined by a monolayer of epithelial cells that acts as a selectively permeable barrier, preventing the passage of harmful intraluminal foreign antigens, flora, and toxins into the circulation while allowing digestion and absorption of essential dietary nutrients along with the transfer of electrolytes and water.Proteins in the tight junctions of intestinal epithelial cells maintain barrier integrity, but barrier dysfunction occurs when these cells are damaged in the setting of infection, burns, shock, or hypoxia (low oxygen levels). The transcription factor HIF-1, a heterodimer composed of the macromolecules HIF-1α and HIF-1β, regulates the adaptive cellular response to hypoxia and the consequent expression of tight junction proteins.Researchers assessed the concentration of HIF-1 heterodimer components in human intestinal Caco-2 cells subjected to hypoxia/reoxygenation (H/R) in vitro. Caco-2 cells, a colon-derived cell line, were cultured under specific conditions to mimic the functional and morphological phenotype of wild-type enterocytes lining the small intestine. These cells were prepared and grown as a monolayer on a collagen-coated membrane.Next, the monolayer was cultured in hypoxic conditions and then exposed to atmospheric oxygen levels (normoxia) for 30, 60, and 120 minutes. Protein levels were quantified using direct enzyme-linked immunosorbent assay (ELISA), in which an antibody linked to a reporter enzyme was utilized to bind and detect expression of the target molecule (analyte) in a sample. When the colorless substrate of the reporter enzyme was added, the enzyme generated a visible colored product that could be quantified based on color intensity.
 Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Adapted from Lei Q, Qiang F, Chao D, et al. Int J Mol Med. 2014;34(6):1629-39.
Which of the following statements about HIF-1 is most likely to be true?At low oxygen levels, HIF-1 is nonfunctional.At low oxygen levels, HIF-1 is functional.At normal oxygen levels, HIF-1 concentration is decreased.At normal oxygen levels, HIF-1 concentration is increased.
A)I and III only
B)I and IV only
C)II and III only
D)II and IV only
The lumen of the human gut is lined by a monolayer of epithelial cells that acts as a selectively permeable barrier, preventing the passage of harmful intraluminal foreign antigens, flora, and toxins into the circulation while allowing digestion and absorption of essential dietary nutrients along with the transfer of electrolytes and water.Proteins in the tight junctions of intestinal epithelial cells maintain barrier integrity, but barrier dysfunction occurs when these cells are damaged in the setting of infection, burns, shock, or hypoxia (low oxygen levels). The transcription factor HIF-1, a heterodimer composed of the macromolecules HIF-1α and HIF-1β, regulates the adaptive cellular response to hypoxia and the consequent expression of tight junction proteins.Researchers assessed the concentration of HIF-1 heterodimer components in human intestinal Caco-2 cells subjected to hypoxia/reoxygenation (H/R) in vitro. Caco-2 cells, a colon-derived cell line, were cultured under specific conditions to mimic the functional and morphological phenotype of wild-type enterocytes lining the small intestine. These cells were prepared and grown as a monolayer on a collagen-coated membrane.Next, the monolayer was cultured in hypoxic conditions and then exposed to atmospheric oxygen levels (normoxia) for 30, 60, and 120 minutes. Protein levels were quantified using direct enzyme-linked immunosorbent assay (ELISA), in which an antibody linked to a reporter enzyme was utilized to bind and detect expression of the target molecule (analyte) in a sample. When the colorless substrate of the reporter enzyme was added, the enzyme generated a visible colored product that could be quantified based on color intensity.
 Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.Adapted from Lei Q, Qiang F, Chao D, et al. Int J Mol Med. 2014;34(6):1629-39.
Which of the following statements about HIF-1 is most likely to be true?At low oxygen levels, HIF-1 is nonfunctional.At low oxygen levels, HIF-1 is functional.At normal oxygen levels, HIF-1 concentration is decreased.At normal oxygen levels, HIF-1 concentration is increased.
A)I and III only
B)I and IV only
C)II and III only
D)II and IV only

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
21
Passage
Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 105 SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.Table 1 Mean anti-SRC titers with standard error
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_2c1d_a3bf_85095183ef29_MD0008_00.jpg) Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_532e_a3bf_2bc230da9462_MD0008_00.jpg) Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?
A)![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f43_d171_a3bf_41d636b7e3ec_MD0008_00.jpg)
B)![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f43_f882_a3bf_e1fa106ae4a1_MD0008_00.jpg)
C)![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f44_1f93_a3bf_3723ecb8b55d_MD0008_00.jpg)
D)![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f44_1f94_a3bf_81e6826eb40a_MD0008_00.jpg)
Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 105 SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.Table 1 Mean anti-SRC titers with standard error
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_2c1d_a3bf_85095183ef29_MD0008_00.jpg) Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_532e_a3bf_2bc230da9462_MD0008_00.jpg) Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?
A)
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f43_d171_a3bf_41d636b7e3ec_MD0008_00.jpg)
B)
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f43_f882_a3bf_e1fa106ae4a1_MD0008_00.jpg)
C)
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f44_1f93_a3bf_3723ecb8b55d_MD0008_00.jpg)
D)
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. According to the results of Experiment 3, which of the following could represent survival curves for F1 and He-J mice?</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f44_1f94_a3bf_81e6826eb40a_MD0008_00.jpg)

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
22
Passage
The Bloom syndrome helicase (BLM) transcript is catalytically inactive in Bloom syndrome (BS), a genetic disorder that leads to genomic instability characterized by excessive somatic recombination events. Research studies have shown that BLM may be involved in DNA synthesis repair of stalled replication forks during replication stress (arrest). To investigate this further, an experiment was conducted using cell lines derived from a patient with BS. One set of cells differed only by BLM status: PSNF5 (BLM+, vector with BLM cDNA transfected) and PSNG13 (BLM−, empty vector transfected). Additional mutations, BLM-K695T and BLM-T99A, were studied in stable transfected cells.The nucleoside analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were used to label and track genomic regions of active replication. Cells were initially treated with IdU for 15 minutes. Subsequently, either 4 mM of hydroxyurea (HU) or 30 µM of aphidicolin (Aph) were used to induce replication stress for 4 hours, after which cells were treated with CldU for 85 minutes to assess replication recovery. Cells were mounted on microscope slides, lysed to isolate chromosome fibers, and then fixed on the slides. Sites of IdU and CldU incorporation into the growing DNA strand were then detected by immunostaining the isolated chromosome fibers with analog-specific, fluorescent-coupled antibodies. Replication activity was visualized via microscopy and measured.
 Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
 Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
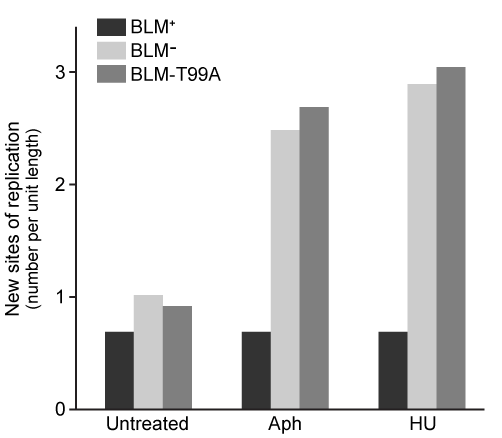 Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Adapted from Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24(3):1279-91.
After the 85-minute period, deoxyribonucleoside triphosphates (dNTPs) are added and DNA synthesis is allowed to continue for 20 minutes. Which of the following processes is most likely to occur during the new replication period? The structure of CldU is shown below.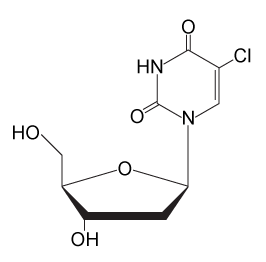
A)CldU and a dNTP molecule would react endergonically to release a pyrophosphate molecule.
B)CldU and a dNTP molecule would react exergonically to form a covalent phosphodiester bond.
C)The 3′ OH group from the last nucleotide of the growing strand would attack the 5′ PO4 group of an incoming dNTP.
D)The 5′ PO4 group from the last nucleotide of the growing strand would attack the 3′ OH group of an incoming dNTP.
The Bloom syndrome helicase (BLM) transcript is catalytically inactive in Bloom syndrome (BS), a genetic disorder that leads to genomic instability characterized by excessive somatic recombination events. Research studies have shown that BLM may be involved in DNA synthesis repair of stalled replication forks during replication stress (arrest). To investigate this further, an experiment was conducted using cell lines derived from a patient with BS. One set of cells differed only by BLM status: PSNF5 (BLM+, vector with BLM cDNA transfected) and PSNG13 (BLM−, empty vector transfected). Additional mutations, BLM-K695T and BLM-T99A, were studied in stable transfected cells.The nucleoside analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were used to label and track genomic regions of active replication. Cells were initially treated with IdU for 15 minutes. Subsequently, either 4 mM of hydroxyurea (HU) or 30 µM of aphidicolin (Aph) were used to induce replication stress for 4 hours, after which cells were treated with CldU for 85 minutes to assess replication recovery. Cells were mounted on microscope slides, lysed to isolate chromosome fibers, and then fixed on the slides. Sites of IdU and CldU incorporation into the growing DNA strand were then detected by immunostaining the isolated chromosome fibers with analog-specific, fluorescent-coupled antibodies. Replication activity was visualized via microscopy and measured.
 Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type) Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)Adapted from Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24(3):1279-91.
After the 85-minute period, deoxyribonucleoside triphosphates (dNTPs) are added and DNA synthesis is allowed to continue for 20 minutes. Which of the following processes is most likely to occur during the new replication period? The structure of CldU is shown below.

A)CldU and a dNTP molecule would react endergonically to release a pyrophosphate molecule.
B)CldU and a dNTP molecule would react exergonically to form a covalent phosphodiester bond.
C)The 3′ OH group from the last nucleotide of the growing strand would attack the 5′ PO4 group of an incoming dNTP.
D)The 5′ PO4 group from the last nucleotide of the growing strand would attack the 3′ OH group of an incoming dNTP.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
23
Passage
Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects.
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. A patient treated for cirrhosis was given vitamin B<sub>6</sub> to stimulate red blood cell production. To counteract the potential negative side effects of this treatment, extract of ginkgo biloba was also administered. What is the most likely role of ginkgo biloba extract in this scenario?</strong> A)It stimulates hemoglobin synthesis B)It increases hemoglobin's oxygen affinity C)It neutralizes reactive oxygen species D)It inhibits blood acidification](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_43f3_a3bf_497266064431_MD0008_00.jpg) Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. A patient treated for cirrhosis was given vitamin B<sub>6</sub> to stimulate red blood cell production. To counteract the potential negative side effects of this treatment, extract of ginkgo biloba was also administered. What is the most likely role of ginkgo biloba extract in this scenario?</strong> A)It stimulates hemoglobin synthesis B)It increases hemoglobin's oxygen affinity C)It neutralizes reactive oxygen species D)It inhibits blood acidification](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_6b04_a3bf_6b4009cc2af4_MD0008_00.jpg) Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
A patient treated for cirrhosis was given vitamin B6 to stimulate red blood cell production. To counteract the potential negative side effects of this treatment, extract of ginkgo biloba was also administered. What is the most likely role of ginkgo biloba extract in this scenario?
A)It stimulates hemoglobin synthesis
B)It increases hemoglobin's oxygen affinity
C)It neutralizes reactive oxygen species
D)It inhibits blood acidification
Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects.
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. A patient treated for cirrhosis was given vitamin B<sub>6</sub> to stimulate red blood cell production. To counteract the potential negative side effects of this treatment, extract of ginkgo biloba was also administered. What is the most likely role of ginkgo biloba extract in this scenario?</strong> A)It stimulates hemoglobin synthesis B)It increases hemoglobin's oxygen affinity C)It neutralizes reactive oxygen species D)It inhibits blood acidification](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_43f3_a3bf_497266064431_MD0008_00.jpg) Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. A patient treated for cirrhosis was given vitamin B<sub>6</sub> to stimulate red blood cell production. To counteract the potential negative side effects of this treatment, extract of ginkgo biloba was also administered. What is the most likely role of ginkgo biloba extract in this scenario?</strong> A)It stimulates hemoglobin synthesis B)It increases hemoglobin's oxygen affinity C)It neutralizes reactive oxygen species D)It inhibits blood acidification](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_6b04_a3bf_6b4009cc2af4_MD0008_00.jpg) Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.A patient treated for cirrhosis was given vitamin B6 to stimulate red blood cell production. To counteract the potential negative side effects of this treatment, extract of ginkgo biloba was also administered. What is the most likely role of ginkgo biloba extract in this scenario?
A)It stimulates hemoglobin synthesis
B)It increases hemoglobin's oxygen affinity
C)It neutralizes reactive oxygen species
D)It inhibits blood acidification

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
24
Passage
Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects.
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Hyperventilation leads to oxygen dissociation curves similar to those seen in patients with cirrhosis when at rest. Based on this information, what physiological change in cirrhotic patients most likely decreases the level of oxygen delivery to tissues?</strong> A)Low carbonic acid and CO<sub>2</sub> in the blood B)Increased excretion of bicarbonate by kidneys C)Higher PO<sub>2</sub> in the lungs of cirrhotic patients D)Lower affinity of hemoglobin for oxygen in tissues](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_43f3_a3bf_497266064431_MD0008_00.jpg) Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Hyperventilation leads to oxygen dissociation curves similar to those seen in patients with cirrhosis when at rest. Based on this information, what physiological change in cirrhotic patients most likely decreases the level of oxygen delivery to tissues?</strong> A)Low carbonic acid and CO<sub>2</sub> in the blood B)Increased excretion of bicarbonate by kidneys C)Higher PO<sub>2</sub> in the lungs of cirrhotic patients D)Lower affinity of hemoglobin for oxygen in tissues](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_6b04_a3bf_6b4009cc2af4_MD0008_00.jpg) Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Hyperventilation leads to oxygen dissociation curves similar to those seen in patients with cirrhosis when at rest. Based on this information, what physiological change in cirrhotic patients most likely decreases the level of oxygen delivery to tissues?
A)Low carbonic acid and CO2 in the blood
B)Increased excretion of bicarbonate by kidneys
C)Higher PO2 in the lungs of cirrhotic patients
D)Lower affinity of hemoglobin for oxygen in tissues
Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects.
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Hyperventilation leads to oxygen dissociation curves similar to those seen in patients with cirrhosis when at rest. Based on this information, what physiological change in cirrhotic patients most likely decreases the level of oxygen delivery to tissues?</strong> A)Low carbonic acid and CO<sub>2</sub> in the blood B)Increased excretion of bicarbonate by kidneys C)Higher PO<sub>2</sub> in the lungs of cirrhotic patients D)Lower affinity of hemoglobin for oxygen in tissues](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_43f3_a3bf_497266064431_MD0008_00.jpg) Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Hyperventilation leads to oxygen dissociation curves similar to those seen in patients with cirrhosis when at rest. Based on this information, what physiological change in cirrhotic patients most likely decreases the level of oxygen delivery to tissues?</strong> A)Low carbonic acid and CO<sub>2</sub> in the blood B)Increased excretion of bicarbonate by kidneys C)Higher PO<sub>2</sub> in the lungs of cirrhotic patients D)Lower affinity of hemoglobin for oxygen in tissues](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_6b04_a3bf_6b4009cc2af4_MD0008_00.jpg) Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.Hyperventilation leads to oxygen dissociation curves similar to those seen in patients with cirrhosis when at rest. Based on this information, what physiological change in cirrhotic patients most likely decreases the level of oxygen delivery to tissues?
A)Low carbonic acid and CO2 in the blood
B)Increased excretion of bicarbonate by kidneys
C)Higher PO2 in the lungs of cirrhotic patients
D)Lower affinity of hemoglobin for oxygen in tissues

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
25
Passage
The Bloom syndrome helicase (BLM) transcript is catalytically inactive in Bloom syndrome (BS), a genetic disorder that leads to genomic instability characterized by excessive somatic recombination events. Research studies have shown that BLM may be involved in DNA synthesis repair of stalled replication forks during replication stress (arrest). To investigate this further, an experiment was conducted using cell lines derived from a patient with BS. One set of cells differed only by BLM status: PSNF5 (BLM+, vector with BLM cDNA transfected) and PSNG13 (BLM−, empty vector transfected). Additional mutations, BLM-K695T and BLM-T99A, were studied in stable transfected cells.The nucleoside analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were used to label and track genomic regions of active replication. Cells were initially treated with IdU for 15 minutes. Subsequently, either 4 mM of hydroxyurea (HU) or 30 µM of aphidicolin (Aph) were used to induce replication stress for 4 hours, after which cells were treated with CldU for 85 minutes to assess replication recovery. Cells were mounted on microscope slides, lysed to isolate chromosome fibers, and then fixed on the slides. Sites of IdU and CldU incorporation into the growing DNA strand were then detected by immunostaining the isolated chromosome fibers with analog-specific, fluorescent-coupled antibodies. Replication activity was visualized via microscopy and measured.
 Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
 Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
 Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Adapted from Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24(3):1279-91.
During HU treatment of PSNF5 cells, replication stress response factors would act at DNA fibers that resemble which rows in Figure 1B?
A)Rows 1 and 2
B)Rows 2 and 5
C)Rows 3 and 4
D)Rows 5 and 6
The Bloom syndrome helicase (BLM) transcript is catalytically inactive in Bloom syndrome (BS), a genetic disorder that leads to genomic instability characterized by excessive somatic recombination events. Research studies have shown that BLM may be involved in DNA synthesis repair of stalled replication forks during replication stress (arrest). To investigate this further, an experiment was conducted using cell lines derived from a patient with BS. One set of cells differed only by BLM status: PSNF5 (BLM+, vector with BLM cDNA transfected) and PSNG13 (BLM−, empty vector transfected). Additional mutations, BLM-K695T and BLM-T99A, were studied in stable transfected cells.The nucleoside analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were used to label and track genomic regions of active replication. Cells were initially treated with IdU for 15 minutes. Subsequently, either 4 mM of hydroxyurea (HU) or 30 µM of aphidicolin (Aph) were used to induce replication stress for 4 hours, after which cells were treated with CldU for 85 minutes to assess replication recovery. Cells were mounted on microscope slides, lysed to isolate chromosome fibers, and then fixed on the slides. Sites of IdU and CldU incorporation into the growing DNA strand were then detected by immunostaining the isolated chromosome fibers with analog-specific, fluorescent-coupled antibodies. Replication activity was visualized via microscopy and measured.
 Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type) Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)Adapted from Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24(3):1279-91.
During HU treatment of PSNF5 cells, replication stress response factors would act at DNA fibers that resemble which rows in Figure 1B?
A)Rows 1 and 2
B)Rows 2 and 5
C)Rows 3 and 4
D)Rows 5 and 6

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
26
Passage
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a bacterium responsible for causing foodborne illnesses. EHEC virulence, or the ability to cause disease, is acquired through quorum sensing, a form of bacterial cell-cell communication dependent on population density. EHEC O157:H7 bacteria monitor population density by detecting changes in the concentration of autoinducer-3 (AI-3), an extracellular signaling molecule secreted by many bacterial species. AI-3 generally acts on neighboring bacteria to activate the genes on the enterocyte effacement (LEE) locus. These genes code for type III secretion proteins (EspA, EspB, EspD, Tir), which promote entry of Shiga-like toxins secreted by EHEC into host intestinal epithelial cells. This ultimately leads to cell death, lesions, and symptoms such as bloody diarrhea and abdominal cramps.It has been reported that a mutation in S-ribosylhomocysteine lyase (LuxS), an enzyme involved in quorum sensing, may result in diminished production of AI-3. However, researchers predict that certain peptide hormones can activate LEE gene transcription in the LuxS. To test this hypothesis, mutant EHEC bacteria deficient in LuxS (LuxS−) were cultured and separately exposed to the hormones epinephrine (E), norepinephrine (NE), gastrin (G), and secretin (S) at 37°C. Total RNA was extracted from the treated bacterial cultures, and LEE mRNA was quantified (Figure 1).
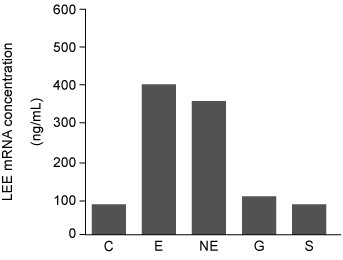 Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
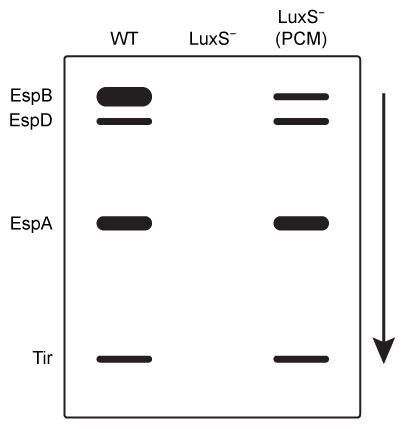 Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
Based on the passage, what best explains the western blot results of type III secretion proteins in LuxS− E. coli cells cultured in PCM?
A)Intestinal epithelial cells secreted secretin and gastrin into the PCM.
B)Type III secretion proteins were produced by intestinal epithelial cells.
C)LuxS− cells underwent a spontaneous reverse mutation that restored normal LuxS function.
D)Compounds released by intestinal epithelial cells into the PCM activated transcription of LEE genes.
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a bacterium responsible for causing foodborne illnesses. EHEC virulence, or the ability to cause disease, is acquired through quorum sensing, a form of bacterial cell-cell communication dependent on population density. EHEC O157:H7 bacteria monitor population density by detecting changes in the concentration of autoinducer-3 (AI-3), an extracellular signaling molecule secreted by many bacterial species. AI-3 generally acts on neighboring bacteria to activate the genes on the enterocyte effacement (LEE) locus. These genes code for type III secretion proteins (EspA, EspB, EspD, Tir), which promote entry of Shiga-like toxins secreted by EHEC into host intestinal epithelial cells. This ultimately leads to cell death, lesions, and symptoms such as bloody diarrhea and abdominal cramps.It has been reported that a mutation in S-ribosylhomocysteine lyase (LuxS), an enzyme involved in quorum sensing, may result in diminished production of AI-3. However, researchers predict that certain peptide hormones can activate LEE gene transcription in the LuxS. To test this hypothesis, mutant EHEC bacteria deficient in LuxS (LuxS−) were cultured and separately exposed to the hormones epinephrine (E), norepinephrine (NE), gastrin (G), and secretin (S) at 37°C. Total RNA was extracted from the treated bacterial cultures, and LEE mRNA was quantified (Figure 1).
 Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria. Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCMBased on the passage, what best explains the western blot results of type III secretion proteins in LuxS− E. coli cells cultured in PCM?
A)Intestinal epithelial cells secreted secretin and gastrin into the PCM.
B)Type III secretion proteins were produced by intestinal epithelial cells.
C)LuxS− cells underwent a spontaneous reverse mutation that restored normal LuxS function.
D)Compounds released by intestinal epithelial cells into the PCM activated transcription of LEE genes.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
27
Passage
The Bloom syndrome helicase (BLM) transcript is catalytically inactive in Bloom syndrome (BS), a genetic disorder that leads to genomic instability characterized by excessive somatic recombination events. Research studies have shown that BLM may be involved in DNA synthesis repair of stalled replication forks during replication stress (arrest). To investigate this further, an experiment was conducted using cell lines derived from a patient with BS. One set of cells differed only by BLM status: PSNF5 (BLM+, vector with BLM cDNA transfected) and PSNG13 (BLM−, empty vector transfected). Additional mutations, BLM-K695T and BLM-T99A, were studied in stable transfected cells.The nucleoside analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were used to label and track genomic regions of active replication. Cells were initially treated with IdU for 15 minutes. Subsequently, either 4 mM of hydroxyurea (HU) or 30 µM of aphidicolin (Aph) were used to induce replication stress for 4 hours, after which cells were treated with CldU for 85 minutes to assess replication recovery. Cells were mounted on microscope slides, lysed to isolate chromosome fibers, and then fixed on the slides. Sites of IdU and CldU incorporation into the growing DNA strand were then detected by immunostaining the isolated chromosome fibers with analog-specific, fluorescent-coupled antibodies. Replication activity was visualized via microscopy and measured.
 Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
 Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
 Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Adapted from Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24(3):1279-91.
All of the following enzymes would be used to generate the vector transfected into PSNF5 cells EXCEPT:
A)Reverse transcriptase
B)RNA polymerase
C)DNA polymerase
D)Restriction enzyme
The Bloom syndrome helicase (BLM) transcript is catalytically inactive in Bloom syndrome (BS), a genetic disorder that leads to genomic instability characterized by excessive somatic recombination events. Research studies have shown that BLM may be involved in DNA synthesis repair of stalled replication forks during replication stress (arrest). To investigate this further, an experiment was conducted using cell lines derived from a patient with BS. One set of cells differed only by BLM status: PSNF5 (BLM+, vector with BLM cDNA transfected) and PSNG13 (BLM−, empty vector transfected). Additional mutations, BLM-K695T and BLM-T99A, were studied in stable transfected cells.The nucleoside analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were used to label and track genomic regions of active replication. Cells were initially treated with IdU for 15 minutes. Subsequently, either 4 mM of hydroxyurea (HU) or 30 µM of aphidicolin (Aph) were used to induce replication stress for 4 hours, after which cells were treated with CldU for 85 minutes to assess replication recovery. Cells were mounted on microscope slides, lysed to isolate chromosome fibers, and then fixed on the slides. Sites of IdU and CldU incorporation into the growing DNA strand were then detected by immunostaining the isolated chromosome fibers with analog-specific, fluorescent-coupled antibodies. Replication activity was visualized via microscopy and measured.
 Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type) Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)Adapted from Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24(3):1279-91.
All of the following enzymes would be used to generate the vector transfected into PSNF5 cells EXCEPT:
A)Reverse transcriptase
B)RNA polymerase
C)DNA polymerase
D)Restriction enzyme

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
28
Passage
The Bloom syndrome helicase (BLM) transcript is catalytically inactive in Bloom syndrome (BS), a genetic disorder that leads to genomic instability characterized by excessive somatic recombination events. Research studies have shown that BLM may be involved in DNA synthesis repair of stalled replication forks during replication stress (arrest). To investigate this further, an experiment was conducted using cell lines derived from a patient with BS. One set of cells differed only by BLM status: PSNF5 (BLM+, vector with BLM cDNA transfected) and PSNG13 (BLM−, empty vector transfected). Additional mutations, BLM-K695T and BLM-T99A, were studied in stable transfected cells.The nucleoside analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were used to label and track genomic regions of active replication. Cells were initially treated with IdU for 15 minutes. Subsequently, either 4 mM of hydroxyurea (HU) or 30 µM of aphidicolin (Aph) were used to induce replication stress for 4 hours, after which cells were treated with CldU for 85 minutes to assess replication recovery. Cells were mounted on microscope slides, lysed to isolate chromosome fibers, and then fixed on the slides. Sites of IdU and CldU incorporation into the growing DNA strand were then detected by immunostaining the isolated chromosome fibers with analog-specific, fluorescent-coupled antibodies. Replication activity was visualized via microscopy and measured.
 Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
 Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
 Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Adapted from Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24(3):1279-91.
The BLM peptide sequence was used to search for similar peptides in various species using an amino acid sequence homology (similarity) database. BLM and other peptides were found to share similar amino acid sequences only in the DNA helicase domains. The results of sequence homology with BLM are shown in the table below.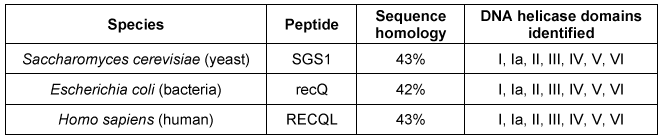 Which statement best explains the role of these peptides during replicative stress in the organisms studied?
Which statement best explains the role of these peptides during replicative stress in the organisms studied?
A)SGS1 would be required to repair replication forks at one origin of replication.
B)recQ would be required to repair replication forks at one origin of replication.
C)BLM would be unable to repair replication forks at multiple origins of replication.
D)RECQL would be unable to repair replication forks at multiple origins of replication.
The Bloom syndrome helicase (BLM) transcript is catalytically inactive in Bloom syndrome (BS), a genetic disorder that leads to genomic instability characterized by excessive somatic recombination events. Research studies have shown that BLM may be involved in DNA synthesis repair of stalled replication forks during replication stress (arrest). To investigate this further, an experiment was conducted using cell lines derived from a patient with BS. One set of cells differed only by BLM status: PSNF5 (BLM+, vector with BLM cDNA transfected) and PSNG13 (BLM−, empty vector transfected). Additional mutations, BLM-K695T and BLM-T99A, were studied in stable transfected cells.The nucleoside analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were used to label and track genomic regions of active replication. Cells were initially treated with IdU for 15 minutes. Subsequently, either 4 mM of hydroxyurea (HU) or 30 µM of aphidicolin (Aph) were used to induce replication stress for 4 hours, after which cells were treated with CldU for 85 minutes to assess replication recovery. Cells were mounted on microscope slides, lysed to isolate chromosome fibers, and then fixed on the slides. Sites of IdU and CldU incorporation into the growing DNA strand were then detected by immunostaining the isolated chromosome fibers with analog-specific, fluorescent-coupled antibodies. Replication activity was visualized via microscopy and measured.
 Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type) Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)Adapted from Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24(3):1279-91.
The BLM peptide sequence was used to search for similar peptides in various species using an amino acid sequence homology (similarity) database. BLM and other peptides were found to share similar amino acid sequences only in the DNA helicase domains. The results of sequence homology with BLM are shown in the table below.
 Which statement best explains the role of these peptides during replicative stress in the organisms studied?
Which statement best explains the role of these peptides during replicative stress in the organisms studied?A)SGS1 would be required to repair replication forks at one origin of replication.
B)recQ would be required to repair replication forks at one origin of replication.
C)BLM would be unable to repair replication forks at multiple origins of replication.
D)RECQL would be unable to repair replication forks at multiple origins of replication.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
29
Passage
The Bloom syndrome helicase (BLM) transcript is catalytically inactive in Bloom syndrome (BS), a genetic disorder that leads to genomic instability characterized by excessive somatic recombination events. Research studies have shown that BLM may be involved in DNA synthesis repair of stalled replication forks during replication stress (arrest). To investigate this further, an experiment was conducted using cell lines derived from a patient with BS. One set of cells differed only by BLM status: PSNF5 (BLM+, vector with BLM cDNA transfected) and PSNG13 (BLM−, empty vector transfected). Additional mutations, BLM-K695T and BLM-T99A, were studied in stable transfected cells.The nucleoside analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were used to label and track genomic regions of active replication. Cells were initially treated with IdU for 15 minutes. Subsequently, either 4 mM of hydroxyurea (HU) or 30 µM of aphidicolin (Aph) were used to induce replication stress for 4 hours, after which cells were treated with CldU for 85 minutes to assess replication recovery. Cells were mounted on microscope slides, lysed to isolate chromosome fibers, and then fixed on the slides. Sites of IdU and CldU incorporation into the growing DNA strand were then detected by immunostaining the isolated chromosome fibers with analog-specific, fluorescent-coupled antibodies. Replication activity was visualized via microscopy and measured.
 Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
 Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
 Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Adapted from Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24(3):1279-91.
What is the most accurate description of Aph-treated cells compared to wild-type cells treated with the same agent?
A)Supercoiling function of DNA topoisomerase is deficient in BLM-T99A cells.
B)Activity of single-stranded DNA binding proteins is increased in BLM-T99A cells.
C)Unwinding activity by DNA helicase is increased in BLM-K695T cells.
D)DNA primer production by primase is unchanged in BLM-K695T cells.
The Bloom syndrome helicase (BLM) transcript is catalytically inactive in Bloom syndrome (BS), a genetic disorder that leads to genomic instability characterized by excessive somatic recombination events. Research studies have shown that BLM may be involved in DNA synthesis repair of stalled replication forks during replication stress (arrest). To investigate this further, an experiment was conducted using cell lines derived from a patient with BS. One set of cells differed only by BLM status: PSNF5 (BLM+, vector with BLM cDNA transfected) and PSNG13 (BLM−, empty vector transfected). Additional mutations, BLM-K695T and BLM-T99A, were studied in stable transfected cells.The nucleoside analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were used to label and track genomic regions of active replication. Cells were initially treated with IdU for 15 minutes. Subsequently, either 4 mM of hydroxyurea (HU) or 30 µM of aphidicolin (Aph) were used to induce replication stress for 4 hours, after which cells were treated with CldU for 85 minutes to assess replication recovery. Cells were mounted on microscope slides, lysed to isolate chromosome fibers, and then fixed on the slides. Sites of IdU and CldU incorporation into the growing DNA strand were then detected by immunostaining the isolated chromosome fibers with analog-specific, fluorescent-coupled antibodies. Replication activity was visualized via microscopy and measured.
 Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type) Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)Adapted from Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24(3):1279-91.
What is the most accurate description of Aph-treated cells compared to wild-type cells treated with the same agent?
A)Supercoiling function of DNA topoisomerase is deficient in BLM-T99A cells.
B)Activity of single-stranded DNA binding proteins is increased in BLM-T99A cells.
C)Unwinding activity by DNA helicase is increased in BLM-K695T cells.
D)DNA primer production by primase is unchanged in BLM-K695T cells.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
30
Passage
Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 105 SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.Table 1 Mean anti-SRC titers with standard error
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. In mice, a brown coat color is a dominant trait. Assuming that the He-N mice in the experiments were homozygous for the white allele and the He-J mice were homozygous for the brown allele, which of the following is true with regard to coat color and LPS response in F1 and F2 mice?</strong> A)F1 mice will produce gametes with equal haplotype frequencies. B)A large fraction of F2 mice exhibiting full response to LPS will be white. C)A large fraction of F1 mice exhibiting full response to LPS will be white. D)Half of F1 mice exhibiting full response to LPS will be brown and the other half will be white.](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_2c1d_a3bf_85095183ef29_MD0008_00.jpg) Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. In mice, a brown coat color is a dominant trait. Assuming that the He-N mice in the experiments were homozygous for the white allele and the He-J mice were homozygous for the brown allele, which of the following is true with regard to coat color and LPS response in F1 and F2 mice?</strong> A)F1 mice will produce gametes with equal haplotype frequencies. B)A large fraction of F2 mice exhibiting full response to LPS will be white. C)A large fraction of F1 mice exhibiting full response to LPS will be white. D)Half of F1 mice exhibiting full response to LPS will be brown and the other half will be white.](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_532e_a3bf_2bc230da9462_MD0008_00.jpg) Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
In mice, a brown coat color is a dominant trait. Assuming that the He-N mice in the experiments were homozygous for the white allele and the He-J mice were homozygous for the brown allele, which of the following is true with regard to coat color and LPS response in F1 and F2 mice?
A)F1 mice will produce gametes with equal haplotype frequencies.
B)A large fraction of F2 mice exhibiting full response to LPS will be white.
C)A large fraction of F1 mice exhibiting full response to LPS will be white.
D)Half of F1 mice exhibiting full response to LPS will be brown and the other half will be white.
Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 105 SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.Table 1 Mean anti-SRC titers with standard error
![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. In mice, a brown coat color is a dominant trait. Assuming that the He-N mice in the experiments were homozygous for the white allele and the He-J mice were homozygous for the brown allele, which of the following is true with regard to coat color and LPS response in F1 and F2 mice?</strong> A)F1 mice will produce gametes with equal haplotype frequencies. B)A large fraction of F2 mice exhibiting full response to LPS will be white. C)A large fraction of F1 mice exhibiting full response to LPS will be white. D)Half of F1 mice exhibiting full response to LPS will be brown and the other half will be white.](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_2c1d_a3bf_85095183ef29_MD0008_00.jpg) Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.
Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus.![<strong>Passage Lipopolysaccharides (LPS) found in gram-negative bacterial cells can elicit a lethal degree of adjuvanticity, or immune activation, in LPS-responsive host organisms. In mice, responses may be modulated by abnormalities in the toll-like receptor (TLR) encoded by the Tlr4 gene closely linked to coat color on Chromosome 4. To investigate the effect of a particular Tlr4 mutation on LPS response, three experiments were performed following a cross between fully responsive He-N mice [tlr4(+)/tlr4(+)] and He-J mice homozygous for the mutation [tlr4(d)/tlr4(d)].Experiment 1Researchers isolated spleen cells for analysis of protein expression and LPS response. In cells isolated from F1 mice, both wild-type and mutant TLRs were distinguishable by protein electrophoresis. Following this first analysis, spleen cells from each cohort were exposed to 2 × 10<sup>5</sup> SRC, an antigen formed by sheep erythrocytes. Cultures were then incubated in the presence or absence of Ph-LPS, with LPS extracted from Escherichia coli K345 cells using a phenol-water mixture. Titers of anti-SRC antibody were recorded on incubation day 5, with the results shown in Table 1.<strong>Table 1</strong> Mean anti-SRC titers with standard error Experiment 2Serum samples were assayed for interferon production 2 hours post LPS injection. Interferon was quantified using a plaque reduction assay on mouse cells challenged with vesicular stomatitis virus. <strong>Figure 1</strong> Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD<sub>50</sub>) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD<sub>50</sub> for F1 mice and a 100-fold increase in LD<sub>50</sub> for He-J mice. In mice, a brown coat color is a dominant trait. Assuming that the He-N mice in the experiments were homozygous for the white allele and the He-J mice were homozygous for the brown allele, which of the following is true with regard to coat color and LPS response in F1 and F2 mice?</strong> A)F1 mice will produce gametes with equal haplotype frequencies. B)A large fraction of F2 mice exhibiting full response to LPS will be white. C)A large fraction of F1 mice exhibiting full response to LPS will be white. D)Half of F1 mice exhibiting full response to LPS will be brown and the other half will be white.](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f3e_532e_a3bf_2bc230da9462_MD0008_00.jpg) Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.
Figure 1 Average serum interferon levels in mice cohortsExperiment 3Mice were injected with varying doses of Ph-LPS and monitored for 4 days. The lethal dose (LD50) was defined as the minimum dose causing death in 50% of injected mice. Compared to He-N mice, results reflected a 10-fold increase in LD50 for F1 mice and a 100-fold increase in LD50 for He-J mice.In mice, a brown coat color is a dominant trait. Assuming that the He-N mice in the experiments were homozygous for the white allele and the He-J mice were homozygous for the brown allele, which of the following is true with regard to coat color and LPS response in F1 and F2 mice?
A)F1 mice will produce gametes with equal haplotype frequencies.
B)A large fraction of F2 mice exhibiting full response to LPS will be white.
C)A large fraction of F1 mice exhibiting full response to LPS will be white.
D)Half of F1 mice exhibiting full response to LPS will be brown and the other half will be white.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
31
Passage
Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure.
![<strong>Passage Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure. <strong>Figure 1</strong> Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.<strong>Table 1</strong> Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8. If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f56_6f2c_a3bf_990c0f0c4c2c_MD0008_00.jpg) Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
![<strong>Passage Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure. <strong>Figure 1</strong> Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.<strong>Table 1</strong> Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8. If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f56_963d_a3bf_dfd0075f45cd_MD0008_00.jpg) Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].
A)![<strong>Passage Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure. <strong>Figure 1</strong> Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.<strong>Table 1</strong> Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8. If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f56_bd4e_a3bf_f34008751516_MD0008_00.jpg)
B)![<strong>Passage Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure. <strong>Figure 1</strong> Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.<strong>Table 1</strong> Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8. If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f56_e45f_a3bf_7fd30bae9411_MD0008_00.jpg)
C)![<strong>Passage Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure. <strong>Figure 1</strong> Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.<strong>Table 1</strong> Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8. If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f56_e460_a3bf_e7ae39b581d5_MD0008_00.jpg)
D)![<strong>Passage Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure. <strong>Figure 1</strong> Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.<strong>Table 1</strong> Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8. If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f57_0b71_a3bf_e34d472781f4_MD0008_00.jpg)
Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure.
![<strong>Passage Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure. <strong>Figure 1</strong> Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.<strong>Table 1</strong> Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8. If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f56_6f2c_a3bf_990c0f0c4c2c_MD0008_00.jpg) Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility
Figure 1 Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.Table 1 Effect of HMGB1 and HMGA1 on H1-GFP Mobility![<strong>Passage Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure. <strong>Figure 1</strong> Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.<strong>Table 1</strong> Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8. If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f56_963d_a3bf_dfd0075f45cd_MD0008_00.jpg) Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.
Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8.If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].
A)
![<strong>Passage Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure. <strong>Figure 1</strong> Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.<strong>Table 1</strong> Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8. If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f56_bd4e_a3bf_f34008751516_MD0008_00.jpg)
B)
![<strong>Passage Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure. <strong>Figure 1</strong> Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.<strong>Table 1</strong> Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8. If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f56_e45f_a3bf_7fd30bae9411_MD0008_00.jpg)
C)
![<strong>Passage Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure. <strong>Figure 1</strong> Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.<strong>Table 1</strong> Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8. If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f56_e460_a3bf_e7ae39b581d5_MD0008_00.jpg)
D)
![<strong>Passage Quantitative fluorescence recovery after photobleaching (FRAP) is used to study the movement of molecules in live cells. During FRAP, the fluorescence of a green fluorescent protein (GFP)-tagged molecule is first measured and then photodestroyed in targeted cell regions. Researchers then assess the time course for fluorescence recovery, which is an indicator of molecular mobility of the GFP-tagged protein into the photodestroyed region. The average time required to recover 80% of the pre-bleach fluorescence of protein histone 1 (H1) fused to GFP (H1-GFP) in the photodestroyed region is given as t80.FRAP was used to analyze the mobility of the architectural H1 alone and in the presence of high-mobility group (HMG) proteins. HMG proteins are dynamic modifiers that have been shown to have opposite effects but similar binding sites on chromatin structure. <strong>Figure 1</strong> Unique chromatin binding motifs of HMG proteinsDuring analysis, HMGA1 and HMGB1 were microinjected into the cytoplasm of mouse embryonic fibroblast cells expressing H1-GFP. FRAP was performed on euchromatin and heterochromatin. Euchromatin and heterochromatin domains were identified as regions weakly and strongly stained by H1-GFP, respectively. The relative intensity of H1-GFP fluorescence in euchromatin and heterochromatin of uninjected and injected cells was assessed, and t80 results are shown in Table 1.<strong>Table 1</strong> Effect of HMGB1 and HMGA1 on H1-GFP Mobility Adapted from Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24(10):4321-8. If HMGA1 were injected into cells expressing HMGB1-GFP instead of H1-GFP, which of the following FRAP analysis graphs would show conclusively that HMGA1 binds the same sites as HMGB1 on chromatin? [The t80 value for HMGB1-GFP is indicated by the arrows].</strong> A) B) C) D)](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f57_0b71_a3bf_e34d472781f4_MD0008_00.jpg)

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
32
Passage
Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects.
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Upregulation of which enzyme could improve oxygen release into the tissues of cirrhotic patients?</strong> A)Phosphoglycerate kinase B)Glyceraldehyde-3-phosphate dehydrogenase C)Bisphosphoglycerate mutase D)Phosphoglycerate mutase](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_43f3_a3bf_497266064431_MD0008_00.jpg) Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Upregulation of which enzyme could improve oxygen release into the tissues of cirrhotic patients?</strong> A)Phosphoglycerate kinase B)Glyceraldehyde-3-phosphate dehydrogenase C)Bisphosphoglycerate mutase D)Phosphoglycerate mutase](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_6b04_a3bf_6b4009cc2af4_MD0008_00.jpg) Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Upregulation of which enzyme could improve oxygen release into the tissues of cirrhotic patients?
A)Phosphoglycerate kinase
B)Glyceraldehyde-3-phosphate dehydrogenase
C)Bisphosphoglycerate mutase
D)Phosphoglycerate mutase
Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects.
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Upregulation of which enzyme could improve oxygen release into the tissues of cirrhotic patients?</strong> A)Phosphoglycerate kinase B)Glyceraldehyde-3-phosphate dehydrogenase C)Bisphosphoglycerate mutase D)Phosphoglycerate mutase](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_43f3_a3bf_497266064431_MD0008_00.jpg) Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Upregulation of which enzyme could improve oxygen release into the tissues of cirrhotic patients?</strong> A)Phosphoglycerate kinase B)Glyceraldehyde-3-phosphate dehydrogenase C)Bisphosphoglycerate mutase D)Phosphoglycerate mutase](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_6b04_a3bf_6b4009cc2af4_MD0008_00.jpg) Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.Upregulation of which enzyme could improve oxygen release into the tissues of cirrhotic patients?
A)Phosphoglycerate kinase
B)Glyceraldehyde-3-phosphate dehydrogenase
C)Bisphosphoglycerate mutase
D)Phosphoglycerate mutase

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
33
Passage
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a bacterium responsible for causing foodborne illnesses. EHEC virulence, or the ability to cause disease, is acquired through quorum sensing, a form of bacterial cell-cell communication dependent on population density. EHEC O157:H7 bacteria monitor population density by detecting changes in the concentration of autoinducer-3 (AI-3), an extracellular signaling molecule secreted by many bacterial species. AI-3 generally acts on neighboring bacteria to activate the genes on the enterocyte effacement (LEE) locus. These genes code for type III secretion proteins (EspA, EspB, EspD, Tir), which promote entry of Shiga-like toxins secreted by EHEC into host intestinal epithelial cells. This ultimately leads to cell death, lesions, and symptoms such as bloody diarrhea and abdominal cramps.It has been reported that a mutation in S-ribosylhomocysteine lyase (LuxS), an enzyme involved in quorum sensing, may result in diminished production of AI-3. However, researchers predict that certain peptide hormones can activate LEE gene transcription in the LuxS. To test this hypothesis, mutant EHEC bacteria deficient in LuxS (LuxS−) were cultured and separately exposed to the hormones epinephrine (E), norepinephrine (NE), gastrin (G), and secretin (S) at 37°C. Total RNA was extracted from the treated bacterial cultures, and LEE mRNA was quantified (Figure 1).
 Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
 Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
E. coli K-12, a normal bacterial strain of the human microbiota, is able to cause disease by inducing enterocyte lesions after it is grown in media with EHEC bacteria that contain the F factor plasmid. What process most likely granted virulence to E. coli K-12?
A)Transformation
B)Transduction
C)Conjugation
D)Transfection
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a bacterium responsible for causing foodborne illnesses. EHEC virulence, or the ability to cause disease, is acquired through quorum sensing, a form of bacterial cell-cell communication dependent on population density. EHEC O157:H7 bacteria monitor population density by detecting changes in the concentration of autoinducer-3 (AI-3), an extracellular signaling molecule secreted by many bacterial species. AI-3 generally acts on neighboring bacteria to activate the genes on the enterocyte effacement (LEE) locus. These genes code for type III secretion proteins (EspA, EspB, EspD, Tir), which promote entry of Shiga-like toxins secreted by EHEC into host intestinal epithelial cells. This ultimately leads to cell death, lesions, and symptoms such as bloody diarrhea and abdominal cramps.It has been reported that a mutation in S-ribosylhomocysteine lyase (LuxS), an enzyme involved in quorum sensing, may result in diminished production of AI-3. However, researchers predict that certain peptide hormones can activate LEE gene transcription in the LuxS. To test this hypothesis, mutant EHEC bacteria deficient in LuxS (LuxS−) were cultured and separately exposed to the hormones epinephrine (E), norepinephrine (NE), gastrin (G), and secretin (S) at 37°C. Total RNA was extracted from the treated bacterial cultures, and LEE mRNA was quantified (Figure 1).
 Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria. Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCME. coli K-12, a normal bacterial strain of the human microbiota, is able to cause disease by inducing enterocyte lesions after it is grown in media with EHEC bacteria that contain the F factor plasmid. What process most likely granted virulence to E. coli K-12?
A)Transformation
B)Transduction
C)Conjugation
D)Transfection

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
34
Passage
Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects.
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Healthy subjects show a shallow increase in oxygen-carrying capacity at 0-20 mm Hg, but a sharp increase at 20-50 mm Hg. Which statements explain this effect?Oxygen binding induces a change from the R state to the T stateThe affinity of hemoglobin for oxygen increases with O<sub>2</sub> pressureHemoglobin exhibits positive binding cooperativityHemoglobin exhibits negative binding cooperativity</strong> A)I and IV B)II and III C)I, II, and III D)I, II, and IV](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_43f3_a3bf_497266064431_MD0008_00.jpg) Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Healthy subjects show a shallow increase in oxygen-carrying capacity at 0-20 mm Hg, but a sharp increase at 20-50 mm Hg. Which statements explain this effect?Oxygen binding induces a change from the R state to the T stateThe affinity of hemoglobin for oxygen increases with O<sub>2</sub> pressureHemoglobin exhibits positive binding cooperativityHemoglobin exhibits negative binding cooperativity</strong> A)I and IV B)II and III C)I, II, and III D)I, II, and IV](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_6b04_a3bf_6b4009cc2af4_MD0008_00.jpg) Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Healthy subjects show a shallow increase in oxygen-carrying capacity at 0-20 mm Hg, but a sharp increase at 20-50 mm Hg. Which statements explain this effect?Oxygen binding induces a change from the R state to the T stateThe affinity of hemoglobin for oxygen increases with O2 pressureHemoglobin exhibits positive binding cooperativityHemoglobin exhibits negative binding cooperativity
A)I and IV
B)II and III
C)I, II, and III
D)I, II, and IV
Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects.
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Healthy subjects show a shallow increase in oxygen-carrying capacity at 0-20 mm Hg, but a sharp increase at 20-50 mm Hg. Which statements explain this effect?Oxygen binding induces a change from the R state to the T stateThe affinity of hemoglobin for oxygen increases with O<sub>2</sub> pressureHemoglobin exhibits positive binding cooperativityHemoglobin exhibits negative binding cooperativity</strong> A)I and IV B)II and III C)I, II, and III D)I, II, and IV](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_43f3_a3bf_497266064431_MD0008_00.jpg) Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Healthy subjects show a shallow increase in oxygen-carrying capacity at 0-20 mm Hg, but a sharp increase at 20-50 mm Hg. Which statements explain this effect?Oxygen binding induces a change from the R state to the T stateThe affinity of hemoglobin for oxygen increases with O<sub>2</sub> pressureHemoglobin exhibits positive binding cooperativityHemoglobin exhibits negative binding cooperativity</strong> A)I and IV B)II and III C)I, II, and III D)I, II, and IV](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_6b04_a3bf_6b4009cc2af4_MD0008_00.jpg) Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.Healthy subjects show a shallow increase in oxygen-carrying capacity at 0-20 mm Hg, but a sharp increase at 20-50 mm Hg. Which statements explain this effect?Oxygen binding induces a change from the R state to the T stateThe affinity of hemoglobin for oxygen increases with O2 pressureHemoglobin exhibits positive binding cooperativityHemoglobin exhibits negative binding cooperativity
A)I and IV
B)II and III
C)I, II, and III
D)I, II, and IV

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
35
Passage
The Bloom syndrome helicase (BLM) transcript is catalytically inactive in Bloom syndrome (BS), a genetic disorder that leads to genomic instability characterized by excessive somatic recombination events. Research studies have shown that BLM may be involved in DNA synthesis repair of stalled replication forks during replication stress (arrest). To investigate this further, an experiment was conducted using cell lines derived from a patient with BS. One set of cells differed only by BLM status: PSNF5 (BLM+, vector with BLM cDNA transfected) and PSNG13 (BLM−, empty vector transfected). Additional mutations, BLM-K695T and BLM-T99A, were studied in stable transfected cells.The nucleoside analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were used to label and track genomic regions of active replication. Cells were initially treated with IdU for 15 minutes. Subsequently, either 4 mM of hydroxyurea (HU) or 30 µM of aphidicolin (Aph) were used to induce replication stress for 4 hours, after which cells were treated with CldU for 85 minutes to assess replication recovery. Cells were mounted on microscope slides, lysed to isolate chromosome fibers, and then fixed on the slides. Sites of IdU and CldU incorporation into the growing DNA strand were then detected by immunostaining the isolated chromosome fibers with analog-specific, fluorescent-coupled antibodies. Replication activity was visualized via microscopy and measured.
 Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
 Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
 Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Adapted from Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24(3):1279-91.
After Aph treatment, replication fork recovery in PSNF5 and PSNG13 cells was analyzed as a function of time. Which of the following conclusions explains the data shown in the graph below?
A)The time at which replication fork recovery was assessed serves to predict BLM expression levels in both cell types.
B)During replication fork recovery, the rates of DNA synthesis and somatic recombination differ in the two cell types.
C)PSNG13 cells are less sensitive to the effects of Aph than PSNF5 cells.
D)Recovery time is dependent on the percentage of replication forks that can be rescued.
The Bloom syndrome helicase (BLM) transcript is catalytically inactive in Bloom syndrome (BS), a genetic disorder that leads to genomic instability characterized by excessive somatic recombination events. Research studies have shown that BLM may be involved in DNA synthesis repair of stalled replication forks during replication stress (arrest). To investigate this further, an experiment was conducted using cell lines derived from a patient with BS. One set of cells differed only by BLM status: PSNF5 (BLM+, vector with BLM cDNA transfected) and PSNG13 (BLM−, empty vector transfected). Additional mutations, BLM-K695T and BLM-T99A, were studied in stable transfected cells.The nucleoside analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were used to label and track genomic regions of active replication. Cells were initially treated with IdU for 15 minutes. Subsequently, either 4 mM of hydroxyurea (HU) or 30 µM of aphidicolin (Aph) were used to induce replication stress for 4 hours, after which cells were treated with CldU for 85 minutes to assess replication recovery. Cells were mounted on microscope slides, lysed to isolate chromosome fibers, and then fixed on the slides. Sites of IdU and CldU incorporation into the growing DNA strand were then detected by immunostaining the isolated chromosome fibers with analog-specific, fluorescent-coupled antibodies. Replication activity was visualized via microscopy and measured.
 Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type) Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)Adapted from Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24(3):1279-91.
After Aph treatment, replication fork recovery in PSNF5 and PSNG13 cells was analyzed as a function of time. Which of the following conclusions explains the data shown in the graph below?

A)The time at which replication fork recovery was assessed serves to predict BLM expression levels in both cell types.
B)During replication fork recovery, the rates of DNA synthesis and somatic recombination differ in the two cell types.
C)PSNG13 cells are less sensitive to the effects of Aph than PSNF5 cells.
D)Recovery time is dependent on the percentage of replication forks that can be rescued.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
36
Passage
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a bacterium responsible for causing foodborne illnesses. EHEC virulence, or the ability to cause disease, is acquired through quorum sensing, a form of bacterial cell-cell communication dependent on population density. EHEC O157:H7 bacteria monitor population density by detecting changes in the concentration of autoinducer-3 (AI-3), an extracellular signaling molecule secreted by many bacterial species. AI-3 generally acts on neighboring bacteria to activate the genes on the enterocyte effacement (LEE) locus. These genes code for type III secretion proteins (EspA, EspB, EspD, Tir), which promote entry of Shiga-like toxins secreted by EHEC into host intestinal epithelial cells. This ultimately leads to cell death, lesions, and symptoms such as bloody diarrhea and abdominal cramps.It has been reported that a mutation in S-ribosylhomocysteine lyase (LuxS), an enzyme involved in quorum sensing, may result in diminished production of AI-3. However, researchers predict that certain peptide hormones can activate LEE gene transcription in the LuxS. To test this hypothesis, mutant EHEC bacteria deficient in LuxS (LuxS−) were cultured and separately exposed to the hormones epinephrine (E), norepinephrine (NE), gastrin (G), and secretin (S) at 37°C. Total RNA was extracted from the treated bacterial cultures, and LEE mRNA was quantified (Figure 1).
 Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
 Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
Which of the following is most likely to be found within the cell membranes of intestinal epithelial cells?
A)Peptidoglycan
B)Cholesterol
C)Cytoskeletal filaments
D)Cellulose
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a bacterium responsible for causing foodborne illnesses. EHEC virulence, or the ability to cause disease, is acquired through quorum sensing, a form of bacterial cell-cell communication dependent on population density. EHEC O157:H7 bacteria monitor population density by detecting changes in the concentration of autoinducer-3 (AI-3), an extracellular signaling molecule secreted by many bacterial species. AI-3 generally acts on neighboring bacteria to activate the genes on the enterocyte effacement (LEE) locus. These genes code for type III secretion proteins (EspA, EspB, EspD, Tir), which promote entry of Shiga-like toxins secreted by EHEC into host intestinal epithelial cells. This ultimately leads to cell death, lesions, and symptoms such as bloody diarrhea and abdominal cramps.It has been reported that a mutation in S-ribosylhomocysteine lyase (LuxS), an enzyme involved in quorum sensing, may result in diminished production of AI-3. However, researchers predict that certain peptide hormones can activate LEE gene transcription in the LuxS. To test this hypothesis, mutant EHEC bacteria deficient in LuxS (LuxS−) were cultured and separately exposed to the hormones epinephrine (E), norepinephrine (NE), gastrin (G), and secretin (S) at 37°C. Total RNA was extracted from the treated bacterial cultures, and LEE mRNA was quantified (Figure 1).
 Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria. Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCMWhich of the following is most likely to be found within the cell membranes of intestinal epithelial cells?
A)Peptidoglycan
B)Cholesterol
C)Cytoskeletal filaments
D)Cellulose

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
37
Passage
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a bacterium responsible for causing foodborne illnesses. EHEC virulence, or the ability to cause disease, is acquired through quorum sensing, a form of bacterial cell-cell communication dependent on population density. EHEC O157:H7 bacteria monitor population density by detecting changes in the concentration of autoinducer-3 (AI-3), an extracellular signaling molecule secreted by many bacterial species. AI-3 generally acts on neighboring bacteria to activate the genes on the enterocyte effacement (LEE) locus. These genes code for type III secretion proteins (EspA, EspB, EspD, Tir), which promote entry of Shiga-like toxins secreted by EHEC into host intestinal epithelial cells. This ultimately leads to cell death, lesions, and symptoms such as bloody diarrhea and abdominal cramps.It has been reported that a mutation in S-ribosylhomocysteine lyase (LuxS), an enzyme involved in quorum sensing, may result in diminished production of AI-3. However, researchers predict that certain peptide hormones can activate LEE gene transcription in the LuxS. To test this hypothesis, mutant EHEC bacteria deficient in LuxS (LuxS−) were cultured and separately exposed to the hormones epinephrine (E), norepinephrine (NE), gastrin (G), and secretin (S) at 37°C. Total RNA was extracted from the treated bacterial cultures, and LEE mRNA was quantified (Figure 1).
 Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
 Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
EHEC is classified as a bacilli bacterium. Given this information, this type of classification is most likely based on the bacterium's:
A)habitat.
B)oxygen dependence.
C)morphology.
D)virulence.
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a bacterium responsible for causing foodborne illnesses. EHEC virulence, or the ability to cause disease, is acquired through quorum sensing, a form of bacterial cell-cell communication dependent on population density. EHEC O157:H7 bacteria monitor population density by detecting changes in the concentration of autoinducer-3 (AI-3), an extracellular signaling molecule secreted by many bacterial species. AI-3 generally acts on neighboring bacteria to activate the genes on the enterocyte effacement (LEE) locus. These genes code for type III secretion proteins (EspA, EspB, EspD, Tir), which promote entry of Shiga-like toxins secreted by EHEC into host intestinal epithelial cells. This ultimately leads to cell death, lesions, and symptoms such as bloody diarrhea and abdominal cramps.It has been reported that a mutation in S-ribosylhomocysteine lyase (LuxS), an enzyme involved in quorum sensing, may result in diminished production of AI-3. However, researchers predict that certain peptide hormones can activate LEE gene transcription in the LuxS. To test this hypothesis, mutant EHEC bacteria deficient in LuxS (LuxS−) were cultured and separately exposed to the hormones epinephrine (E), norepinephrine (NE), gastrin (G), and secretin (S) at 37°C. Total RNA was extracted from the treated bacterial cultures, and LEE mRNA was quantified (Figure 1).
 Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria. Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCMEHEC is classified as a bacilli bacterium. Given this information, this type of classification is most likely based on the bacterium's:
A)habitat.
B)oxygen dependence.
C)morphology.
D)virulence.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
38
Passage
The lumen of the human gut is lined by a monolayer of epithelial cells that acts as a selectively permeable barrier, preventing the passage of harmful intraluminal foreign antigens, flora, and toxins into the circulation while allowing digestion and absorption of essential dietary nutrients along with the transfer of electrolytes and water.Proteins in the tight junctions of intestinal epithelial cells maintain barrier integrity, but barrier dysfunction occurs when these cells are damaged in the setting of infection, burns, shock, or hypoxia (low oxygen levels). The transcription factor HIF-1, a heterodimer composed of the macromolecules HIF-1α and HIF-1β, regulates the adaptive cellular response to hypoxia and the consequent expression of tight junction proteins.Researchers assessed the concentration of HIF-1 heterodimer components in human intestinal Caco-2 cells subjected to hypoxia/reoxygenation (H/R) in vitro. Caco-2 cells, a colon-derived cell line, were cultured under specific conditions to mimic the functional and morphological phenotype of wild-type enterocytes lining the small intestine. These cells were prepared and grown as a monolayer on a collagen-coated membrane.Next, the monolayer was cultured in hypoxic conditions and then exposed to atmospheric oxygen levels (normoxia) for 30, 60, and 120 minutes. Protein levels were quantified using direct enzyme-linked immunosorbent assay (ELISA), in which an antibody linked to a reporter enzyme was utilized to bind and detect expression of the target molecule (analyte) in a sample. When the colorless substrate of the reporter enzyme was added, the enzyme generated a visible colored product that could be quantified based on color intensity.
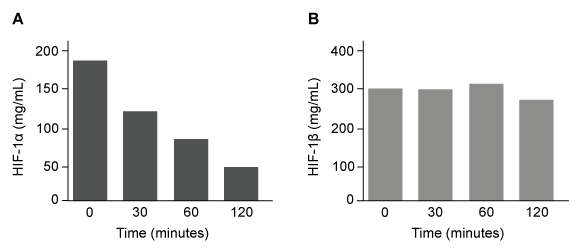 Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Adapted from Lei Q, Qiang F, Chao D, et al. Int J Mol Med. 2014;34(6):1629-39.
A scientist claims that ELISA, although it can accurately detect the concentration of an analyte in a sample, can generate faulty results under certain experimental settings. Inaccurate quantification of an analyte such as HIF-1α using ELISA would most likely result from all of the following EXCEPT:
A)supplementing the sample with a denaturing agent during the initial sample preparation.
B)adding an amount of substrate that is disproportionate to the amount of analyte.
C)detecting unbound antibodies in the sample after quantifying a color change.
D)identifying a color change proportional to analyte concentration after adding the substrate.
The lumen of the human gut is lined by a monolayer of epithelial cells that acts as a selectively permeable barrier, preventing the passage of harmful intraluminal foreign antigens, flora, and toxins into the circulation while allowing digestion and absorption of essential dietary nutrients along with the transfer of electrolytes and water.Proteins in the tight junctions of intestinal epithelial cells maintain barrier integrity, but barrier dysfunction occurs when these cells are damaged in the setting of infection, burns, shock, or hypoxia (low oxygen levels). The transcription factor HIF-1, a heterodimer composed of the macromolecules HIF-1α and HIF-1β, regulates the adaptive cellular response to hypoxia and the consequent expression of tight junction proteins.Researchers assessed the concentration of HIF-1 heterodimer components in human intestinal Caco-2 cells subjected to hypoxia/reoxygenation (H/R) in vitro. Caco-2 cells, a colon-derived cell line, were cultured under specific conditions to mimic the functional and morphological phenotype of wild-type enterocytes lining the small intestine. These cells were prepared and grown as a monolayer on a collagen-coated membrane.Next, the monolayer was cultured in hypoxic conditions and then exposed to atmospheric oxygen levels (normoxia) for 30, 60, and 120 minutes. Protein levels were quantified using direct enzyme-linked immunosorbent assay (ELISA), in which an antibody linked to a reporter enzyme was utilized to bind and detect expression of the target molecule (analyte) in a sample. When the colorless substrate of the reporter enzyme was added, the enzyme generated a visible colored product that could be quantified based on color intensity.
 Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.
Figure 1 Concentration of (A) HIF-1α and (B) HIF-1β in Caco-2 cells subjected to H/R (Note: 0 minutes = cells cultured in hypoxic conditions)Emodin, an anthraquinone compound that prevents hypoxia-induced epithelial cell disruption, was used in conjunction with a chemical HIF-1α inhibitor (HIF-1α-I) to treat Caco-2 cells in a separate experiment. Transepithelial electrical resistance (TEER) was assessed as a measure of barrier function, with higher TEER values indicating a more intact epithelial cell barrier. HIF-1α-I was found to block emodin's protective effect on epithelial barrier integrity.Adapted from Lei Q, Qiang F, Chao D, et al. Int J Mol Med. 2014;34(6):1629-39.
A scientist claims that ELISA, although it can accurately detect the concentration of an analyte in a sample, can generate faulty results under certain experimental settings. Inaccurate quantification of an analyte such as HIF-1α using ELISA would most likely result from all of the following EXCEPT:
A)supplementing the sample with a denaturing agent during the initial sample preparation.
B)adding an amount of substrate that is disproportionate to the amount of analyte.
C)detecting unbound antibodies in the sample after quantifying a color change.
D)identifying a color change proportional to analyte concentration after adding the substrate.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
39
Passage
Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects.
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. The effect of anaerobic exercise on the oxygen saturation curve in healthy patients is shown in Figure 1. Which of the following is involved in the change in the oxygen dissociation curve?</strong> A)CO<sub>2</sub> production decreases in contracting muscle B)Exercise reduces the PO<sub>2</sub> of oxygen in muscles C)Hemoglobin binds oxygen tightly during exercise D)Lactic acid production increases during exercise](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_43f3_a3bf_497266064431_MD0008_00.jpg) Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. The effect of anaerobic exercise on the oxygen saturation curve in healthy patients is shown in Figure 1. Which of the following is involved in the change in the oxygen dissociation curve?</strong> A)CO<sub>2</sub> production decreases in contracting muscle B)Exercise reduces the PO<sub>2</sub> of oxygen in muscles C)Hemoglobin binds oxygen tightly during exercise D)Lactic acid production increases during exercise](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_6b04_a3bf_6b4009cc2af4_MD0008_00.jpg) Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
The effect of anaerobic exercise on the oxygen saturation curve in healthy patients is shown in Figure 1. Which of the following is involved in the change in the oxygen dissociation curve?
A)CO2 production decreases in contracting muscle
B)Exercise reduces the PO2 of oxygen in muscles
C)Hemoglobin binds oxygen tightly during exercise
D)Lactic acid production increases during exercise
Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects.
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. The effect of anaerobic exercise on the oxygen saturation curve in healthy patients is shown in Figure 1. Which of the following is involved in the change in the oxygen dissociation curve?</strong> A)CO<sub>2</sub> production decreases in contracting muscle B)Exercise reduces the PO<sub>2</sub> of oxygen in muscles C)Hemoglobin binds oxygen tightly during exercise D)Lactic acid production increases during exercise](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_43f3_a3bf_497266064431_MD0008_00.jpg) Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. The effect of anaerobic exercise on the oxygen saturation curve in healthy patients is shown in Figure 1. Which of the following is involved in the change in the oxygen dissociation curve?</strong> A)CO<sub>2</sub> production decreases in contracting muscle B)Exercise reduces the PO<sub>2</sub> of oxygen in muscles C)Hemoglobin binds oxygen tightly during exercise D)Lactic acid production increases during exercise](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_6b04_a3bf_6b4009cc2af4_MD0008_00.jpg) Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.The effect of anaerobic exercise on the oxygen saturation curve in healthy patients is shown in Figure 1. Which of the following is involved in the change in the oxygen dissociation curve?
A)CO2 production decreases in contracting muscle
B)Exercise reduces the PO2 of oxygen in muscles
C)Hemoglobin binds oxygen tightly during exercise
D)Lactic acid production increases during exercise

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
40
Passage
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a bacterium responsible for causing foodborne illnesses. EHEC virulence, or the ability to cause disease, is acquired through quorum sensing, a form of bacterial cell-cell communication dependent on population density. EHEC O157:H7 bacteria monitor population density by detecting changes in the concentration of autoinducer-3 (AI-3), an extracellular signaling molecule secreted by many bacterial species. AI-3 generally acts on neighboring bacteria to activate the genes on the enterocyte effacement (LEE) locus. These genes code for type III secretion proteins (EspA, EspB, EspD, Tir), which promote entry of Shiga-like toxins secreted by EHEC into host intestinal epithelial cells. This ultimately leads to cell death, lesions, and symptoms such as bloody diarrhea and abdominal cramps.It has been reported that a mutation in S-ribosylhomocysteine lyase (LuxS), an enzyme involved in quorum sensing, may result in diminished production of AI-3. However, researchers predict that certain peptide hormones can activate LEE gene transcription in the LuxS. To test this hypothesis, mutant EHEC bacteria deficient in LuxS (LuxS−) were cultured and separately exposed to the hormones epinephrine (E), norepinephrine (NE), gastrin (G), and secretin (S) at 37°C. Total RNA was extracted from the treated bacterial cultures, and LEE mRNA was quantified (Figure 1).
 Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
 Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
Which proteins identified in Figure 2 have the shortest amino acid sequence and the greatest concentration in WT cells, respectively?
A)EspA, Tir
B)Tir, EspB
C)EspB, EspA
D)EspB, EspD
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a bacterium responsible for causing foodborne illnesses. EHEC virulence, or the ability to cause disease, is acquired through quorum sensing, a form of bacterial cell-cell communication dependent on population density. EHEC O157:H7 bacteria monitor population density by detecting changes in the concentration of autoinducer-3 (AI-3), an extracellular signaling molecule secreted by many bacterial species. AI-3 generally acts on neighboring bacteria to activate the genes on the enterocyte effacement (LEE) locus. These genes code for type III secretion proteins (EspA, EspB, EspD, Tir), which promote entry of Shiga-like toxins secreted by EHEC into host intestinal epithelial cells. This ultimately leads to cell death, lesions, and symptoms such as bloody diarrhea and abdominal cramps.It has been reported that a mutation in S-ribosylhomocysteine lyase (LuxS), an enzyme involved in quorum sensing, may result in diminished production of AI-3. However, researchers predict that certain peptide hormones can activate LEE gene transcription in the LuxS. To test this hypothesis, mutant EHEC bacteria deficient in LuxS (LuxS−) were cultured and separately exposed to the hormones epinephrine (E), norepinephrine (NE), gastrin (G), and secretin (S) at 37°C. Total RNA was extracted from the treated bacterial cultures, and LEE mRNA was quantified (Figure 1).
 Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria.
Figure 1 Concentration of LEE mRNA in mutant LuxS− bacteria supplemented with purified peptide hormones (Note: C = control.)Western blot was performed to compare the presence and concentration of type III secretion proteins from wild-type (WT), LuxS−, and LuxS− cultured in preconditioned media (PCM). PCM was generated by culturing uninfected intestinal epithelial cells in growth media for 24 hours. After 24 hours, the cells were removed and the PCM was used to supplement the growth of LuxS− bacteria. Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCM
Figure 2 Western blot results of type III secretion proteins isolated from WT, mutant LuxS−, and LuxS− in PCMWhich proteins identified in Figure 2 have the shortest amino acid sequence and the greatest concentration in WT cells, respectively?
A)EspA, Tir
B)Tir, EspB
C)EspB, EspA
D)EspB, EspD

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
41
Passage
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
 Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
 Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Adapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
What conclusion can be made from the results shown in Figure 3?
A)siRNA only binds the mRNA transcripts of proteins that facilitate IRES-ribosome binding.
B)siRNA signals for the degradation of proteins that facilitate elongation of tau v2 proteins.
C)eIF4e mRNA is translated via a cap-independent process.
D)eEF2K mRNA is translated via a cap-dependent process.
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene. Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3). Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNAAdapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
What conclusion can be made from the results shown in Figure 3?
A)siRNA only binds the mRNA transcripts of proteins that facilitate IRES-ribosome binding.
B)siRNA signals for the degradation of proteins that facilitate elongation of tau v2 proteins.
C)eIF4e mRNA is translated via a cap-independent process.
D)eEF2K mRNA is translated via a cap-dependent process.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
42
Passage
Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects.
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Individuals with glucose-6-phosphate dehydrogenase (G6PDH) deficiencies are more susceptible to oxidative stress than others. If a patient with cirrhosis also suffered from G6PDH deficiency, which vitamin could cause the most severe reduction in that patient's hematocrit?</strong> A)Vitamin B<sub>1</sub> B)Vitamin B<sub>3</sub> C)Vitamin B<sub>6</sub> D)Vitamin B<sub>12</sub>](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_43f3_a3bf_497266064431_MD0008_00.jpg) Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Individuals with glucose-6-phosphate dehydrogenase (G6PDH) deficiencies are more susceptible to oxidative stress than others. If a patient with cirrhosis also suffered from G6PDH deficiency, which vitamin could cause the most severe reduction in that patient's hematocrit?</strong> A)Vitamin B<sub>1</sub> B)Vitamin B<sub>3</sub> C)Vitamin B<sub>6</sub> D)Vitamin B<sub>12</sub>](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_6b04_a3bf_6b4009cc2af4_MD0008_00.jpg) Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Individuals with glucose-6-phosphate dehydrogenase (G6PDH) deficiencies are more susceptible to oxidative stress than others. If a patient with cirrhosis also suffered from G6PDH deficiency, which vitamin could cause the most severe reduction in that patient's hematocrit?
A)Vitamin B1
B)Vitamin B3
C)Vitamin B6
D)Vitamin B12
Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects.
![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Individuals with glucose-6-phosphate dehydrogenase (G6PDH) deficiencies are more susceptible to oxidative stress than others. If a patient with cirrhosis also suffered from G6PDH deficiency, which vitamin could cause the most severe reduction in that patient's hematocrit?</strong> A)Vitamin B<sub>1</sub> B)Vitamin B<sub>3</sub> C)Vitamin B<sub>6</sub> D)Vitamin B<sub>12</sub>](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_43f3_a3bf_497266064431_MD0008_00.jpg) Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm
Figure 1 Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.Table 1 Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm![<strong>Passage Hematological anomalies have been observed in patients with prolonged liver damage. For example, patients with chronic cirrhosis (scarring of the liver) often develop anemia and other blood disorders. Hemoglobin is the metalloprotein within red blood cells that is responsible for the oxygen-carrying capacity of blood, and its function may be affected by liver damage. The poor prognosis of patients with cirrhosis and anemia has stimulated further study of hemoglobin function and oxygen saturation.Blood samples from both cirrhotic patients and healthy individuals at rest and after exercise were exposed to varying partial pressures of oxygen. Measurement of oxyhemoglobin dissociation curves (ODCs [Figure 1]) from the blood samples revealed significant functional differences in the hemoglobin from cirrhotic patients relative to control subjects. <strong>Figure 1</strong> Oxyhemoglobin dissociation curve for cirrhotic patients (CP) at rest and for healthy controls (HC) at rest and after 20 minutes of exerciseVitamins are considered nontoxic supplements and are frequently provided to anemic patients to increase red blood cell production. They may therefore have therapeutic benefits for cirrhotic patients. However, some studies indicate that excessive vitamin consumption may lead to oxidative stress that could induce anemia through hemolysis (red blood cell rupturing). Hemoglobin strongly absorbs light at 577 nm, and researchers conducted hemolysis assays by spinning down red blood cells and measuring absorbance (Abs) of the supernatant at 577 nm to determine how much hemoglobin was released. The assay was carried out in the presence of four vitamins to determine their potential as oxidizing agents, and results are shown in Table 1.<strong>Table 1</strong> Hemolysis of Red Blood Cells from Healthy Donors Treated With Vitamins [2 mM] for 30 Minutes, Measured by Absorbance at 577 nm Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45. Individuals with glucose-6-phosphate dehydrogenase (G6PDH) deficiencies are more susceptible to oxidative stress than others. If a patient with cirrhosis also suffered from G6PDH deficiency, which vitamin could cause the most severe reduction in that patient's hematocrit?</strong> A)Vitamin B<sub>1</sub> B)Vitamin B<sub>3</sub> C)Vitamin B<sub>6</sub> D)Vitamin B<sub>12</sub>](https://d2lvgg3v3hfg70.cloudfront.net/MD0008/11ecba2d_0f58_6b04_a3bf_6b4009cc2af4_MD0008_00.jpg) Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.
Adapted from Clerbaux T, Detry B, Geubel A, et al. The oxyhemoglobin dissociation curve in liver cirrhosis. Chest. 2006;129(2):438-45.Individuals with glucose-6-phosphate dehydrogenase (G6PDH) deficiencies are more susceptible to oxidative stress than others. If a patient with cirrhosis also suffered from G6PDH deficiency, which vitamin could cause the most severe reduction in that patient's hematocrit?
A)Vitamin B1
B)Vitamin B3
C)Vitamin B6
D)Vitamin B12

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
43
Passage
The lac operon encodes the proteins that metabolize lactose and is a key biochemical feature that differentiates between members of the Enterobacteriaceae family. For example, the severely virulent Salmonella typhimurium can be distinguished from its less virulent relative Escherichia coli by the absence of a lac operon. Researchers propose that the evolutionary loss of the lac operon may contribute to S. typhimurium's ability to invade epithelial cells.To test the hypothesis, wild-type (WT) S. typhimurium cells were transformed with plasmids encoding components of the lac operon (Figure 1): β-galactosidase (lacZ+), lactose permease (lacY+), transacetylase (lacA+), or all three (lac+). Each plasmid also contained genes for the lac repressor (lacI), which can inhibit lac operon transcription in the absence of lactose; the cAMP receptor protein (crp), which induces lac expression in low glucose; and a chloramphenicol resistance gene (cat).
 Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
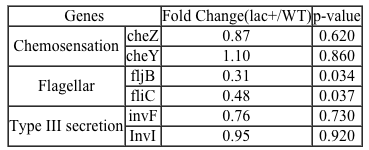
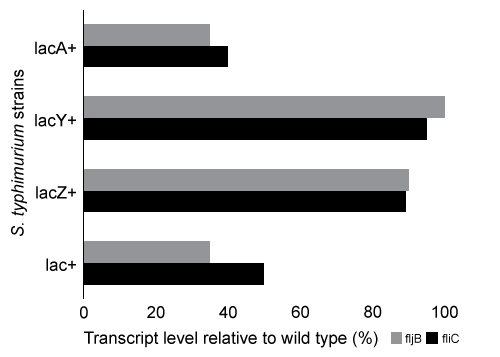 Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Adapted from Jiang L, Ni Z, Wang L, Feng L, Liu B. Loss of the lac operon contributes to Salmonella invasion of epithelial cells through derepression of flagellar synthesis. Curr Microbiol. 2015;70(3):315-23.
Three antibiotic disks were placed on a bacterial culture. One disk contained gentamicin (G), one contained chloramphenicol (C), and one contained ampicillin (A). Based on this information, which of the following cultures represents a strain of Salmonella that has been transformed with the plasmid shown in Figure 1?
A)
B)
C)
D)
The lac operon encodes the proteins that metabolize lactose and is a key biochemical feature that differentiates between members of the Enterobacteriaceae family. For example, the severely virulent Salmonella typhimurium can be distinguished from its less virulent relative Escherichia coli by the absence of a lac operon. Researchers propose that the evolutionary loss of the lac operon may contribute to S. typhimurium's ability to invade epithelial cells.To test the hypothesis, wild-type (WT) S. typhimurium cells were transformed with plasmids encoding components of the lac operon (Figure 1): β-galactosidase (lacZ+), lactose permease (lacY+), transacetylase (lacA+), or all three (lac+). Each plasmid also contained genes for the lac repressor (lacI), which can inhibit lac operon transcription in the absence of lactose; the cAMP receptor protein (crp), which induces lac expression in low glucose; and a chloramphenicol resistance gene (cat).
 Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
 Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.Adapted from Jiang L, Ni Z, Wang L, Feng L, Liu B. Loss of the lac operon contributes to Salmonella invasion of epithelial cells through derepression of flagellar synthesis. Curr Microbiol. 2015;70(3):315-23.
Three antibiotic disks were placed on a bacterial culture. One disk contained gentamicin (G), one contained chloramphenicol (C), and one contained ampicillin (A). Based on this information, which of the following cultures represents a strain of Salmonella that has been transformed with the plasmid shown in Figure 1?
A)

B)

C)

D)


Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
44
Passage
The lac operon encodes the proteins that metabolize lactose and is a key biochemical feature that differentiates between members of the Enterobacteriaceae family. For example, the severely virulent Salmonella typhimurium can be distinguished from its less virulent relative Escherichia coli by the absence of a lac operon. Researchers propose that the evolutionary loss of the lac operon may contribute to S. typhimurium's ability to invade epithelial cells.To test the hypothesis, wild-type (WT) S. typhimurium cells were transformed with plasmids encoding components of the lac operon (Figure 1): β-galactosidase (lacZ+), lactose permease (lacY+), transacetylase (lacA+), or all three (lac+). Each plasmid also contained genes for the lac repressor (lacI), which can inhibit lac operon transcription in the absence of lactose; the cAMP receptor protein (crp), which induces lac expression in low glucose; and a chloramphenicol resistance gene (cat).
 Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT

 Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Adapted from Jiang L, Ni Z, Wang L, Feng L, Liu B. Loss of the lac operon contributes to Salmonella invasion of epithelial cells through derepression of flagellar synthesis. Curr Microbiol. 2015;70(3):315-23.
Based on Figure 2, which of the following would most likely reestablish the virulence of S. typhimurium transformed with lac+?
A)Excising the lacA sequence from the plasmid
B)Stimulating transcription of the lac operon
C)Removing the mRNA that encodes β-galactosidase
D)Increasing expression of lactose permease
The lac operon encodes the proteins that metabolize lactose and is a key biochemical feature that differentiates between members of the Enterobacteriaceae family. For example, the severely virulent Salmonella typhimurium can be distinguished from its less virulent relative Escherichia coli by the absence of a lac operon. Researchers propose that the evolutionary loss of the lac operon may contribute to S. typhimurium's ability to invade epithelial cells.To test the hypothesis, wild-type (WT) S. typhimurium cells were transformed with plasmids encoding components of the lac operon (Figure 1): β-galactosidase (lacZ+), lactose permease (lacY+), transacetylase (lacA+), or all three (lac+). Each plasmid also contained genes for the lac repressor (lacI), which can inhibit lac operon transcription in the absence of lactose; the cAMP receptor protein (crp), which induces lac expression in low glucose; and a chloramphenicol resistance gene (cat).
 Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
 Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.Adapted from Jiang L, Ni Z, Wang L, Feng L, Liu B. Loss of the lac operon contributes to Salmonella invasion of epithelial cells through derepression of flagellar synthesis. Curr Microbiol. 2015;70(3):315-23.
Based on Figure 2, which of the following would most likely reestablish the virulence of S. typhimurium transformed with lac+?
A)Excising the lacA sequence from the plasmid
B)Stimulating transcription of the lac operon
C)Removing the mRNA that encodes β-galactosidase
D)Increasing expression of lactose permease

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
45
Passage
Overexpression of the miR-17~92 gene cluster occurs in many human cancers involving c-Myc, a gene coding for a transcription factor that regulates expression of proteins such as p21, an inhibitor of cell cycle progression. miR-17~92 encodes six distinct microRNAs (miRNAs) that can be divided into four families based on sequence identity at positions 2-7 (Table 1).Table 1 Sequence Identities of miRNA Families Encoded by the mir-17∼92 Gene Cluster
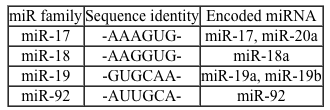 Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
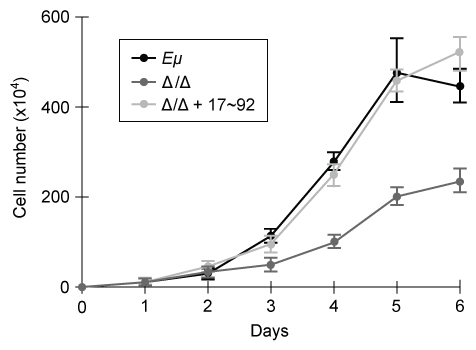 Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
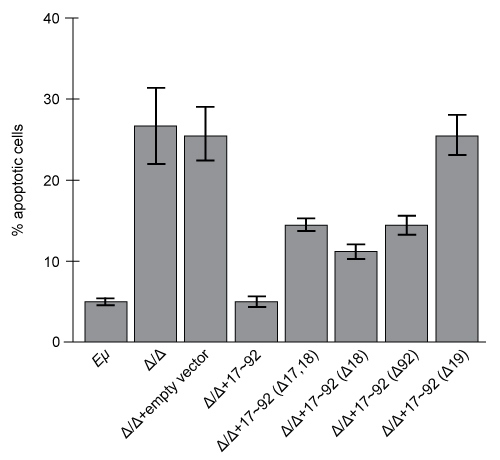 Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Adapted from Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806-11.
As described in the passage, verification that the transduced cells expressed the desired miRNA was an essential experimental step to ensure that:
A)lack of expression of one or more miRNA encoded by the miR-17∼92 cluster did not affect the rate of apoptosis in tested cells.
B)transduction of tested cells with mutant miR-17∼92 alleles did not induce the development of B-cell lymphoma.
C)deletion of one or more miRNA encoded by the miR-17∼92 cluster did not affect expression of the remaining miRNAs encoded by the constructs.
D)expression of one or more mutant miR-17∼92 alleles did not provide the transduced cells with a selective growth advantage.
Overexpression of the miR-17~92 gene cluster occurs in many human cancers involving c-Myc, a gene coding for a transcription factor that regulates expression of proteins such as p21, an inhibitor of cell cycle progression. miR-17~92 encodes six distinct microRNAs (miRNAs) that can be divided into four families based on sequence identity at positions 2-7 (Table 1).Table 1 Sequence Identities of miRNA Families Encoded by the mir-17∼92 Gene Cluster
 Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92). Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2). Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)Adapted from Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806-11.
As described in the passage, verification that the transduced cells expressed the desired miRNA was an essential experimental step to ensure that:
A)lack of expression of one or more miRNA encoded by the miR-17∼92 cluster did not affect the rate of apoptosis in tested cells.
B)transduction of tested cells with mutant miR-17∼92 alleles did not induce the development of B-cell lymphoma.
C)deletion of one or more miRNA encoded by the miR-17∼92 cluster did not affect expression of the remaining miRNAs encoded by the constructs.
D)expression of one or more mutant miR-17∼92 alleles did not provide the transduced cells with a selective growth advantage.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
46
Passage
Overexpression of the miR-17~92 gene cluster occurs in many human cancers involving c-Myc, a gene coding for a transcription factor that regulates expression of proteins such as p21, an inhibitor of cell cycle progression. miR-17~92 encodes six distinct microRNAs (miRNAs) that can be divided into four families based on sequence identity at positions 2-7 (Table 1).Table 1 Sequence Identities of miRNA Families Encoded by the mir-17∼92 Gene Cluster
 Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
 Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
 Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Adapted from Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806-11.
Based on Figure 2, which family from the miR-17~92 gene cluster is expected to contribute the most to cancer progression?
A)miR-17
B)miR-18
C)miR-19
D)miR-92
Overexpression of the miR-17~92 gene cluster occurs in many human cancers involving c-Myc, a gene coding for a transcription factor that regulates expression of proteins such as p21, an inhibitor of cell cycle progression. miR-17~92 encodes six distinct microRNAs (miRNAs) that can be divided into four families based on sequence identity at positions 2-7 (Table 1).Table 1 Sequence Identities of miRNA Families Encoded by the mir-17∼92 Gene Cluster
 Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92). Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2). Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)Adapted from Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806-11.
Based on Figure 2, which family from the miR-17~92 gene cluster is expected to contribute the most to cancer progression?
A)miR-17
B)miR-18
C)miR-19
D)miR-92

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
47
Passage
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
 Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
 Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Adapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
While analyzing potential therapeutic strategies for AD, researchers would most likely pursue which of the following approaches?
A)Phosphatase upregulation
B)Kinase upregulation
C)Isomerase downregulation
D)Synthase downregulation
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene. Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3). Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNAAdapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
While analyzing potential therapeutic strategies for AD, researchers would most likely pursue which of the following approaches?
A)Phosphatase upregulation
B)Kinase upregulation
C)Isomerase downregulation
D)Synthase downregulation

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
48
Passage
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
 Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
 Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Adapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
Type 1 diabetes mellitus (T1DM) differs from T2DM in that affected patients stop producing insulin at a young age. Which of the following is most likely impaired in T1DM?Exocrine function of pancreatic beta cellsEndocrine function of pancreatic beta cellsExocrine function of pancreatic alpha cellsEndocrine function of pancreatic alpha cells
A)I only
B)II only
C)I and II only
D)III and IV only
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene. Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3). Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNAAdapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
Type 1 diabetes mellitus (T1DM) differs from T2DM in that affected patients stop producing insulin at a young age. Which of the following is most likely impaired in T1DM?Exocrine function of pancreatic beta cellsEndocrine function of pancreatic beta cellsExocrine function of pancreatic alpha cellsEndocrine function of pancreatic alpha cells
A)I only
B)II only
C)I and II only
D)III and IV only

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
49
Passage
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
 Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
 Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Adapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
The data in Figure 2 suggest that ribosomes interact with the:
A)IRES within Construct 2 only.
B)5′ cap of Construct 1 only.
C)5′ cap of Construct 2 or the IRES within Constructs 1 and 2.
D)IRES within Construct 1 or the 5′ caps of Constructs 1 and 2.
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene. Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3). Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNAAdapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
The data in Figure 2 suggest that ribosomes interact with the:
A)IRES within Construct 2 only.
B)5′ cap of Construct 1 only.
C)5′ cap of Construct 2 or the IRES within Constructs 1 and 2.
D)IRES within Construct 1 or the 5′ caps of Constructs 1 and 2.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
50
Passage
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
 Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
 Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Adapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
The translation of tau mRNA would most likely involve:an enzyme with the ability to hydrolyze peptide bonds.tRNA molecules carrying amino acids corresponding to codons on the mRNA molecule.a ribosomal complex capable of reading the target mRNA in a 3′ to 5′ direction.
A)I only
B)II only
C)I and II only
D)I and III only
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene. Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3). Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNAAdapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
The translation of tau mRNA would most likely involve:an enzyme with the ability to hydrolyze peptide bonds.tRNA molecules carrying amino acids corresponding to codons on the mRNA molecule.a ribosomal complex capable of reading the target mRNA in a 3′ to 5′ direction.
A)I only
B)II only
C)I and II only
D)I and III only

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
51
Passage
Overexpression of the miR-17~92 gene cluster occurs in many human cancers involving c-Myc, a gene coding for a transcription factor that regulates expression of proteins such as p21, an inhibitor of cell cycle progression. miR-17~92 encodes six distinct microRNAs (miRNAs) that can be divided into four families based on sequence identity at positions 2-7 (Table 1).Table 1 Sequence Identities of miRNA Families Encoded by the mir-17∼92 Gene Cluster
 Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
 Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
 Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Adapted from Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806-11.
Based on the passage, what conclusions can be made about c-Myc and the miR-17∼92 gene cluster?c-Myc induces miR-17∼92 expression.miR-17~92 induces c-Myc expression.miR-17~92 suppresses apoptosis.
A)I only
B)I and II only
C)I and III only
D)I, II, and III only
Overexpression of the miR-17~92 gene cluster occurs in many human cancers involving c-Myc, a gene coding for a transcription factor that regulates expression of proteins such as p21, an inhibitor of cell cycle progression. miR-17~92 encodes six distinct microRNAs (miRNAs) that can be divided into four families based on sequence identity at positions 2-7 (Table 1).Table 1 Sequence Identities of miRNA Families Encoded by the mir-17∼92 Gene Cluster
 Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92). Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2). Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)Adapted from Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806-11.
Based on the passage, what conclusions can be made about c-Myc and the miR-17∼92 gene cluster?c-Myc induces miR-17∼92 expression.miR-17~92 induces c-Myc expression.miR-17~92 suppresses apoptosis.
A)I only
B)I and II only
C)I and III only
D)I, II, and III only

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
52
Passage
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
 Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
 Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Adapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
The cytotoxic protein X.23A induces conformational changes in the large ribosomal subunit, reducing its catalytic activity. Treating neuronal AD mice cells that overexpress tau protein with X.23A would most likely interfere with the binding of the:
A)70S ribosomes to the nucleolus.
B)80S ribosomes to the rough endoplasmic reticulum.
C)50S ribosomal subunit to the smooth endoplasmic reticulum.
D)40S ribosomal subunit to the Golgi body.
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene. Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3). Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNAAdapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
The cytotoxic protein X.23A induces conformational changes in the large ribosomal subunit, reducing its catalytic activity. Treating neuronal AD mice cells that overexpress tau protein with X.23A would most likely interfere with the binding of the:
A)70S ribosomes to the nucleolus.
B)80S ribosomes to the rough endoplasmic reticulum.
C)50S ribosomal subunit to the smooth endoplasmic reticulum.
D)40S ribosomal subunit to the Golgi body.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
53
Passage
Overexpression of the miR-17~92 gene cluster occurs in many human cancers involving c-Myc, a gene coding for a transcription factor that regulates expression of proteins such as p21, an inhibitor of cell cycle progression. miR-17~92 encodes six distinct microRNAs (miRNAs) that can be divided into four families based on sequence identity at positions 2-7 (Table 1).Table 1 Sequence Identities of miRNA Families Encoded by the mir-17∼92 Gene Cluster
 Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
 Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
 Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Adapted from Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806-11.
If changes in p21 expression contribute to the development of B-cell lymphoma in Eμ-Myc mice, what is the expected result of real-time PCR analysis of c-Myc and p21 expression in wild-type (wt) and Eμ-Myc (Eμ) mice? (Note: mRNA levels are standardized to the expression of ubiquitin (Ub), which is not regulated by c-Myc.)
A)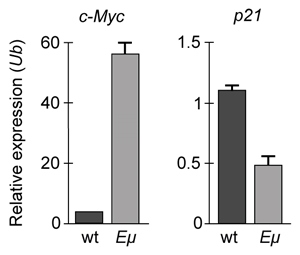
B)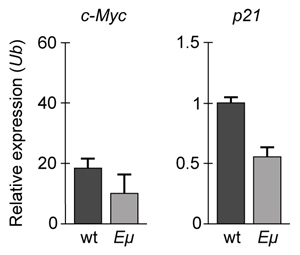
C)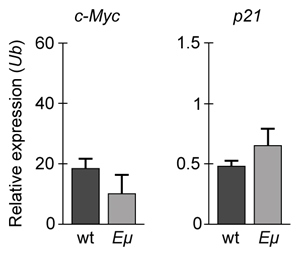
D)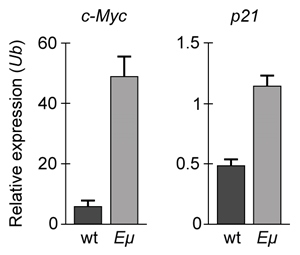
Overexpression of the miR-17~92 gene cluster occurs in many human cancers involving c-Myc, a gene coding for a transcription factor that regulates expression of proteins such as p21, an inhibitor of cell cycle progression. miR-17~92 encodes six distinct microRNAs (miRNAs) that can be divided into four families based on sequence identity at positions 2-7 (Table 1).Table 1 Sequence Identities of miRNA Families Encoded by the mir-17∼92 Gene Cluster
 Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92). Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2). Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)Adapted from Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806-11.
If changes in p21 expression contribute to the development of B-cell lymphoma in Eμ-Myc mice, what is the expected result of real-time PCR analysis of c-Myc and p21 expression in wild-type (wt) and Eμ-Myc (Eμ) mice? (Note: mRNA levels are standardized to the expression of ubiquitin (Ub), which is not regulated by c-Myc.)
A)

B)

C)

D)


Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
54
Passage
The lac operon encodes the proteins that metabolize lactose and is a key biochemical feature that differentiates between members of the Enterobacteriaceae family. For example, the severely virulent Salmonella typhimurium can be distinguished from its less virulent relative Escherichia coli by the absence of a lac operon. Researchers propose that the evolutionary loss of the lac operon may contribute to S. typhimurium's ability to invade epithelial cells.To test the hypothesis, wild-type (WT) S. typhimurium cells were transformed with plasmids encoding components of the lac operon (Figure 1): β-galactosidase (lacZ+), lactose permease (lacY+), transacetylase (lacA+), or all three (lac+). Each plasmid also contained genes for the lac repressor (lacI), which can inhibit lac operon transcription in the absence of lactose; the cAMP receptor protein (crp), which induces lac expression in low glucose; and a chloramphenicol resistance gene (cat).
 Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT

 Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Adapted from Jiang L, Ni Z, Wang L, Feng L, Liu B. Loss of the lac operon contributes to Salmonella invasion of epithelial cells through derepression of flagellar synthesis. Curr Microbiol. 2015;70(3):315-23.
The reduced virulence of lac+ S. typhimurium has been correlated with an inability to generate the torque that turns the flagellum. Which structure in the flagellum is most likely being affected by lac genes?
A)Hook
B)Filament
C)Basal body
D)Cell membrane
The lac operon encodes the proteins that metabolize lactose and is a key biochemical feature that differentiates between members of the Enterobacteriaceae family. For example, the severely virulent Salmonella typhimurium can be distinguished from its less virulent relative Escherichia coli by the absence of a lac operon. Researchers propose that the evolutionary loss of the lac operon may contribute to S. typhimurium's ability to invade epithelial cells.To test the hypothesis, wild-type (WT) S. typhimurium cells were transformed with plasmids encoding components of the lac operon (Figure 1): β-galactosidase (lacZ+), lactose permease (lacY+), transacetylase (lacA+), or all three (lac+). Each plasmid also contained genes for the lac repressor (lacI), which can inhibit lac operon transcription in the absence of lactose; the cAMP receptor protein (crp), which induces lac expression in low glucose; and a chloramphenicol resistance gene (cat).
 Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
 Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.Adapted from Jiang L, Ni Z, Wang L, Feng L, Liu B. Loss of the lac operon contributes to Salmonella invasion of epithelial cells through derepression of flagellar synthesis. Curr Microbiol. 2015;70(3):315-23.
The reduced virulence of lac+ S. typhimurium has been correlated with an inability to generate the torque that turns the flagellum. Which structure in the flagellum is most likely being affected by lac genes?
A)Hook
B)Filament
C)Basal body
D)Cell membrane

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
55
Passage
Overexpression of the miR-17~92 gene cluster occurs in many human cancers involving c-Myc, a gene coding for a transcription factor that regulates expression of proteins such as p21, an inhibitor of cell cycle progression. miR-17~92 encodes six distinct microRNAs (miRNAs) that can be divided into four families based on sequence identity at positions 2-7 (Table 1).Table 1 Sequence Identities of miRNA Families Encoded by the mir-17∼92 Gene Cluster
 Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
 Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
 Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Adapted from Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806-11.
When expressed, the most likely function of the products generated by the miR-17~92 gene cluster is to:
A)assist with the primary enzymatic functions of the ribosome.
B)facilitate the processing of pre-mRNA in the cell nucleus.
C)interfere with gene expression by binding transcripts with complementary nucleotide sequences.
D)prevent small nuclear ribonucleoproteins from forming a large RNA-protein complex.
Overexpression of the miR-17~92 gene cluster occurs in many human cancers involving c-Myc, a gene coding for a transcription factor that regulates expression of proteins such as p21, an inhibitor of cell cycle progression. miR-17~92 encodes six distinct microRNAs (miRNAs) that can be divided into four families based on sequence identity at positions 2-7 (Table 1).Table 1 Sequence Identities of miRNA Families Encoded by the mir-17∼92 Gene Cluster
 Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92).
Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ), which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ). As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92). Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2).
Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation)Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2). Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)Adapted from Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806-11.
When expressed, the most likely function of the products generated by the miR-17~92 gene cluster is to:
A)assist with the primary enzymatic functions of the ribosome.
B)facilitate the processing of pre-mRNA in the cell nucleus.
C)interfere with gene expression by binding transcripts with complementary nucleotide sequences.
D)prevent small nuclear ribonucleoproteins from forming a large RNA-protein complex.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
56
Passage
The lac operon encodes the proteins that metabolize lactose and is a key biochemical feature that differentiates between members of the Enterobacteriaceae family. For example, the severely virulent Salmonella typhimurium can be distinguished from its less virulent relative Escherichia coli by the absence of a lac operon. Researchers propose that the evolutionary loss of the lac operon may contribute to S. typhimurium's ability to invade epithelial cells.To test the hypothesis, wild-type (WT) S. typhimurium cells were transformed with plasmids encoding components of the lac operon (Figure 1): β-galactosidase (lacZ+), lactose permease (lacY+), transacetylase (lacA+), or all three (lac+). Each plasmid also contained genes for the lac repressor (lacI), which can inhibit lac operon transcription in the absence of lactose; the cAMP receptor protein (crp), which induces lac expression in low glucose; and a chloramphenicol resistance gene (cat).
 Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT

 Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Adapted from Jiang L, Ni Z, Wang L, Feng L, Liu B. Loss of the lac operon contributes to Salmonella invasion of epithelial cells through derepression of flagellar synthesis. Curr Microbiol. 2015;70(3):315-23.
Which table shows the expected survival rates of mice infected with WT or lac+ S. typhimurium after being fed a glucose-rich (GR) or a lactose-rich (LR) diet for several weeks?
A)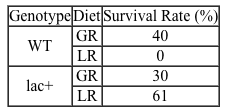
B)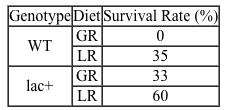
C)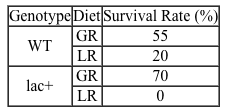
D)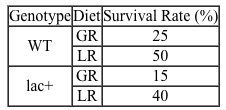
The lac operon encodes the proteins that metabolize lactose and is a key biochemical feature that differentiates between members of the Enterobacteriaceae family. For example, the severely virulent Salmonella typhimurium can be distinguished from its less virulent relative Escherichia coli by the absence of a lac operon. Researchers propose that the evolutionary loss of the lac operon may contribute to S. typhimurium's ability to invade epithelial cells.To test the hypothesis, wild-type (WT) S. typhimurium cells were transformed with plasmids encoding components of the lac operon (Figure 1): β-galactosidase (lacZ+), lactose permease (lacY+), transacetylase (lacA+), or all three (lac+). Each plasmid also contained genes for the lac repressor (lacI), which can inhibit lac operon transcription in the absence of lactose; the cAMP receptor protein (crp), which induces lac expression in low glucose; and a chloramphenicol resistance gene (cat).
 Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
 Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.Adapted from Jiang L, Ni Z, Wang L, Feng L, Liu B. Loss of the lac operon contributes to Salmonella invasion of epithelial cells through derepression of flagellar synthesis. Curr Microbiol. 2015;70(3):315-23.
Which table shows the expected survival rates of mice infected with WT or lac+ S. typhimurium after being fed a glucose-rich (GR) or a lactose-rich (LR) diet for several weeks?
A)

B)

C)

D)


Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
57
Passage
The lac operon encodes the proteins that metabolize lactose and is a key biochemical feature that differentiates between members of the Enterobacteriaceae family. For example, the severely virulent Salmonella typhimurium can be distinguished from its less virulent relative Escherichia coli by the absence of a lac operon. Researchers propose that the evolutionary loss of the lac operon may contribute to S. typhimurium's ability to invade epithelial cells.To test the hypothesis, wild-type (WT) S. typhimurium cells were transformed with plasmids encoding components of the lac operon (Figure 1): β-galactosidase (lacZ+), lactose permease (lacY+), transacetylase (lacA+), or all three (lac+). Each plasmid also contained genes for the lac repressor (lacI), which can inhibit lac operon transcription in the absence of lactose; the cAMP receptor protein (crp), which induces lac expression in low glucose; and a chloramphenicol resistance gene (cat).
 Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT

 Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Adapted from Jiang L, Ni Z, Wang L, Feng L, Liu B. Loss of the lac operon contributes to Salmonella invasion of epithelial cells through derepression of flagellar synthesis. Curr Microbiol. 2015;70(3):315-23.
Which of the following genes would be the most advantageous for lac+ S. typhimurium bacteria in glucose-rich media?
A)cat
B)lacI
C)lacZ
D)crp
The lac operon encodes the proteins that metabolize lactose and is a key biochemical feature that differentiates between members of the Enterobacteriaceae family. For example, the severely virulent Salmonella typhimurium can be distinguished from its less virulent relative Escherichia coli by the absence of a lac operon. Researchers propose that the evolutionary loss of the lac operon may contribute to S. typhimurium's ability to invade epithelial cells.To test the hypothesis, wild-type (WT) S. typhimurium cells were transformed with plasmids encoding components of the lac operon (Figure 1): β-galactosidase (lacZ+), lactose permease (lacY+), transacetylase (lacA+), or all three (lac+). Each plasmid also contained genes for the lac repressor (lacI), which can inhibit lac operon transcription in the absence of lactose; the cAMP receptor protein (crp), which induces lac expression in low glucose; and a chloramphenicol resistance gene (cat).
 Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
 Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.Adapted from Jiang L, Ni Z, Wang L, Feng L, Liu B. Loss of the lac operon contributes to Salmonella invasion of epithelial cells through derepression of flagellar synthesis. Curr Microbiol. 2015;70(3):315-23.
Which of the following genes would be the most advantageous for lac+ S. typhimurium bacteria in glucose-rich media?
A)cat
B)lacI
C)lacZ
D)crp

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
58
Passage
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
 Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
 Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Adapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
Intraperitoneal injection of insulin would cause the peptide hormone to enter the bloodstream and exert its effect on target cells by:
A)acting as a second messenger.
B)diffusing through the plasma membrane.
C)binding an extracellular receptor.
D)disassociating from a carrier protein.
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene. Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3). Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNAAdapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
Intraperitoneal injection of insulin would cause the peptide hormone to enter the bloodstream and exert its effect on target cells by:
A)acting as a second messenger.
B)diffusing through the plasma membrane.
C)binding an extracellular receptor.
D)disassociating from a carrier protein.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
59
Passage
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
 Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
 Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Adapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
Assume gene expression of tau v1 and v2 are synchronized in AD mice cells, and tau mRNA levels are analyzed. Of the following, which technique would researchers use to identify the tau mRNA isoform with the longer half-life in the cytoplasm of AD mice cells?
A)RT-PCR at a single time point
B)Western blot at a single time point
C)RT-PCR at varying time points
D)Western blot at varying time points
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene. Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3). Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNAAdapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
Assume gene expression of tau v1 and v2 are synchronized in AD mice cells, and tau mRNA levels are analyzed. Of the following, which technique would researchers use to identify the tau mRNA isoform with the longer half-life in the cytoplasm of AD mice cells?
A)RT-PCR at a single time point
B)Western blot at a single time point
C)RT-PCR at varying time points
D)Western blot at varying time points

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
60
Passage
The lac operon encodes the proteins that metabolize lactose and is a key biochemical feature that differentiates between members of the Enterobacteriaceae family. For example, the severely virulent Salmonella typhimurium can be distinguished from its less virulent relative Escherichia coli by the absence of a lac operon. Researchers propose that the evolutionary loss of the lac operon may contribute to S. typhimurium's ability to invade epithelial cells.To test the hypothesis, wild-type (WT) S. typhimurium cells were transformed with plasmids encoding components of the lac operon (Figure 1): β-galactosidase (lacZ+), lactose permease (lacY+), transacetylase (lacA+), or all three (lac+). Each plasmid also contained genes for the lac repressor (lacI), which can inhibit lac operon transcription in the absence of lactose; the cAMP receptor protein (crp), which induces lac expression in low glucose; and a chloramphenicol resistance gene (cat).
 Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT

 Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Adapted from Jiang L, Ni Z, Wang L, Feng L, Liu B. Loss of the lac operon contributes to Salmonella invasion of epithelial cells through derepression of flagellar synthesis. Curr Microbiol. 2015;70(3):315-23.
Which of the following conclusions regarding virulence of lac+ S. typhimurium is most accurate?
A)It cannot detect the presence of epithelial cells.
B)It lacks the type III secretion proteins needed for invasion.
C)It infects epithelial cells without using type III secretion proteins.
D)It senses epithelial cells but cannot move toward them.
The lac operon encodes the proteins that metabolize lactose and is a key biochemical feature that differentiates between members of the Enterobacteriaceae family. For example, the severely virulent Salmonella typhimurium can be distinguished from its less virulent relative Escherichia coli by the absence of a lac operon. Researchers propose that the evolutionary loss of the lac operon may contribute to S. typhimurium's ability to invade epithelial cells.To test the hypothesis, wild-type (WT) S. typhimurium cells were transformed with plasmids encoding components of the lac operon (Figure 1): β-galactosidase (lacZ+), lactose permease (lacY+), transacetylase (lacA+), or all three (lac+). Each plasmid also contained genes for the lac repressor (lacI), which can inhibit lac operon transcription in the absence of lactose; the cAMP receptor protein (crp), which induces lac expression in low glucose; and a chloramphenicol resistance gene (cat).
 Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
Figure 1 Plasmid PYM-LACZYA (lac+), used to transform WT S. typhimurium. Similar plasmids containing only one of the three lacZ, lacY, or lacA genes were also used.Virulence assays were performed in mice by inoculating with WT or lac+ S. typhimurium. The assays confirmed that lac+ bacteria were less virulent than their WT counterparts, and that both WT and lac+ bacteria were more virulent in the presence of glucose. Subsequently, several genes related to invasion were analyzed for their expression levels in the presence or absence of lac plasmids (Table 1; Figure 2).Table 1 Transcription Levels of Invasion-Associated Genes in lac+ S. typhimurium Relative to WT
 Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.
Figure 2 Transcript concentrations of two S. typhimurium flagellar genes, fljB and fliC, in bacteria transformed with lacZ+, lacY+, lacA+, or lac+ plasmids. Concentrations are expressed as a percentage of WT levels.Adapted from Jiang L, Ni Z, Wang L, Feng L, Liu B. Loss of the lac operon contributes to Salmonella invasion of epithelial cells through derepression of flagellar synthesis. Curr Microbiol. 2015;70(3):315-23.
Which of the following conclusions regarding virulence of lac+ S. typhimurium is most accurate?
A)It cannot detect the presence of epithelial cells.
B)It lacks the type III secretion proteins needed for invasion.
C)It infects epithelial cells without using type III secretion proteins.
D)It senses epithelial cells but cannot move toward them.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
61
Passage
Obstructive respiratory illnesses are characterized by an increased difficulty exhaling much of the air in the lungs. Asthma and chronic obstructive pulmonary disease (COPD) are two of the most common obstructive respiratory illnesses. Asthma is due to allergen hypersensitivity and can be identified by temporary airway inflammation, which causes the bronchioles to narrow. The decreased airway diameter increases the resistance to airflow. Subsequently, air becomes "trapped" in the lungs, restricting the amount of air that can be moved in each breath.COPD refers to a spectrum of disorders and can be a result of emphysema, the irreversible destruction of pulmonary tissue. The breakdown of elastic proteins in the lungs results in a loss of pulmonary resiliency and chronic lung hyperinflation. Because the walls between individual alveoli are broken down, the alveolar sacs that remain are larger.Lung diseases can be diagnosed and monitored using spirometry. A spirometer measures the flow rate and volume of inhaled and exhaled air. In one pulmonary test, a patient is asked to exhale forcibly and completely into a spirometer after maximum inhalation. This test is done to measure the patient's forced expiratory volume in one second (FEV1) and forced vital capacity (FVC), the total amount of air exhaled in a single breath.
 Figure 1 Forced expiratory spirogram of a healthy individual
Figure 1 Forced expiratory spirogram of a healthy individual
Adapted from T. Barreiro, "An Approach to Interpreting Spirometry." American Family Physician. ©2004 AAFP.
According to the passage, which of the following changes would be expected in a patient with signs of emphysema?
A)Narrowing of the trachea
B)Less efficient gas exchange
C)Weakened diaphragm muscle
D)Overstretched pharyngeal tissues
Obstructive respiratory illnesses are characterized by an increased difficulty exhaling much of the air in the lungs. Asthma and chronic obstructive pulmonary disease (COPD) are two of the most common obstructive respiratory illnesses. Asthma is due to allergen hypersensitivity and can be identified by temporary airway inflammation, which causes the bronchioles to narrow. The decreased airway diameter increases the resistance to airflow. Subsequently, air becomes "trapped" in the lungs, restricting the amount of air that can be moved in each breath.COPD refers to a spectrum of disorders and can be a result of emphysema, the irreversible destruction of pulmonary tissue. The breakdown of elastic proteins in the lungs results in a loss of pulmonary resiliency and chronic lung hyperinflation. Because the walls between individual alveoli are broken down, the alveolar sacs that remain are larger.Lung diseases can be diagnosed and monitored using spirometry. A spirometer measures the flow rate and volume of inhaled and exhaled air. In one pulmonary test, a patient is asked to exhale forcibly and completely into a spirometer after maximum inhalation. This test is done to measure the patient's forced expiratory volume in one second (FEV1) and forced vital capacity (FVC), the total amount of air exhaled in a single breath.
 Figure 1 Forced expiratory spirogram of a healthy individual
Figure 1 Forced expiratory spirogram of a healthy individualAdapted from T. Barreiro, "An Approach to Interpreting Spirometry." American Family Physician. ©2004 AAFP.
According to the passage, which of the following changes would be expected in a patient with signs of emphysema?
A)Narrowing of the trachea
B)Less efficient gas exchange
C)Weakened diaphragm muscle
D)Overstretched pharyngeal tissues

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
62
Passage
Mitochondria are thought to have been independent bacterial organisms that were engulfed and integrated into eukaryotic cells approximately two billion years ago. Most of the mitochondrial genome was lost or transferred into the large central genome of the eukaryotic nucleus, leaving only a residual genome within each mitochondrion.Because multiple mitochondria are found in the cell, mitochondrial DNA (mtDNA) mutations result in a condition known as heteroplasmy, or the intracellular mixture of wild-type mtDNA and mutant mtDNA. Just as the cellular ratio of wild-type to mutant mtDNA varies, the overall mtDNA content within a tissue or organ is also highly variable. For this reason, disease-causing mutant mtDNA manifests phenotypically only when the majority of mitochondria within a given tissue express the deleterious allele. In addition, the variability in mtDNA content makes it possible for individuals with the same mitochondrial mutation to display drastically different clinical symptoms.To investigate how heteroplasmy influences disease manifestation, researchers analyzed an A to G substitution at nucleotide 3243 in the tRNAleu gene of mtDNA. This mutation has been associated with hereditary hearing loss, diabetes mellitus, and a syndrome characterized by mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes.Family members who were confirmed to have the A3243G mutation were tested using an audiometry examination to assess hearing impairment, as shown in Table 1. The subjects carried no other genetic mutations known to cause hearing loss.Table 1 Audiometry Results From Family Members with the A3243G Mutation
 The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
 Figure 1 PAGE visualization of PCR products following ApaI digestion
Figure 1 PAGE visualization of PCR products following ApaI digestion
Adapted from Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc Biol Sci. 2012;279(1734):1865-72.
What conclusion can be drawn from the results shown in Figure 1?
A)The samples were analyzed under denaturing conditions.
B)The samples were analyzed under reducing conditions.
C)The samples were analyzed using a native gel.
D)The samples were analyzed using a stable pH gradient.
Mitochondria are thought to have been independent bacterial organisms that were engulfed and integrated into eukaryotic cells approximately two billion years ago. Most of the mitochondrial genome was lost or transferred into the large central genome of the eukaryotic nucleus, leaving only a residual genome within each mitochondrion.Because multiple mitochondria are found in the cell, mitochondrial DNA (mtDNA) mutations result in a condition known as heteroplasmy, or the intracellular mixture of wild-type mtDNA and mutant mtDNA. Just as the cellular ratio of wild-type to mutant mtDNA varies, the overall mtDNA content within a tissue or organ is also highly variable. For this reason, disease-causing mutant mtDNA manifests phenotypically only when the majority of mitochondria within a given tissue express the deleterious allele. In addition, the variability in mtDNA content makes it possible for individuals with the same mitochondrial mutation to display drastically different clinical symptoms.To investigate how heteroplasmy influences disease manifestation, researchers analyzed an A to G substitution at nucleotide 3243 in the tRNAleu gene of mtDNA. This mutation has been associated with hereditary hearing loss, diabetes mellitus, and a syndrome characterized by mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes.Family members who were confirmed to have the A3243G mutation were tested using an audiometry examination to assess hearing impairment, as shown in Table 1. The subjects carried no other genetic mutations known to cause hearing loss.Table 1 Audiometry Results From Family Members with the A3243G Mutation
 The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI. Figure 1 PAGE visualization of PCR products following ApaI digestion
Figure 1 PAGE visualization of PCR products following ApaI digestionAdapted from Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc Biol Sci. 2012;279(1734):1865-72.
What conclusion can be drawn from the results shown in Figure 1?
A)The samples were analyzed under denaturing conditions.
B)The samples were analyzed under reducing conditions.
C)The samples were analyzed using a native gel.
D)The samples were analyzed using a stable pH gradient.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
63
Passage
Obstructive respiratory illnesses are characterized by an increased difficulty exhaling much of the air in the lungs. Asthma and chronic obstructive pulmonary disease (COPD) are two of the most common obstructive respiratory illnesses. Asthma is due to allergen hypersensitivity and can be identified by temporary airway inflammation, which causes the bronchioles to narrow. The decreased airway diameter increases the resistance to airflow. Subsequently, air becomes "trapped" in the lungs, restricting the amount of air that can be moved in each breath.COPD refers to a spectrum of disorders and can be a result of emphysema, the irreversible destruction of pulmonary tissue. The breakdown of elastic proteins in the lungs results in a loss of pulmonary resiliency and chronic lung hyperinflation. Because the walls between individual alveoli are broken down, the alveolar sacs that remain are larger.Lung diseases can be diagnosed and monitored using spirometry. A spirometer measures the flow rate and volume of inhaled and exhaled air. In one pulmonary test, a patient is asked to exhale forcibly and completely into a spirometer after maximum inhalation. This test is done to measure the patient's forced expiratory volume in one second (FEV1) and forced vital capacity (FVC), the total amount of air exhaled in a single breath.
 Figure 1 Forced expiratory spirogram of a healthy individual
Figure 1 Forced expiratory spirogram of a healthy individual
Adapted from T. Barreiro, "An Approach to Interpreting Spirometry." American Family Physician. ©2004 AAFP.
Which of the following FEV1 spirograms would be expected in a patient with asthma?
A)
B)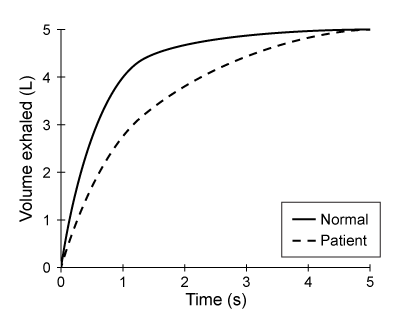
C)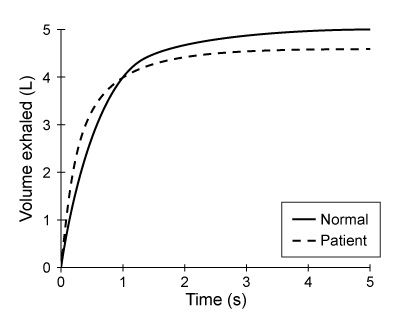
D)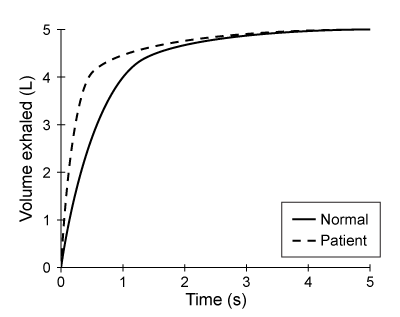
Obstructive respiratory illnesses are characterized by an increased difficulty exhaling much of the air in the lungs. Asthma and chronic obstructive pulmonary disease (COPD) are two of the most common obstructive respiratory illnesses. Asthma is due to allergen hypersensitivity and can be identified by temporary airway inflammation, which causes the bronchioles to narrow. The decreased airway diameter increases the resistance to airflow. Subsequently, air becomes "trapped" in the lungs, restricting the amount of air that can be moved in each breath.COPD refers to a spectrum of disorders and can be a result of emphysema, the irreversible destruction of pulmonary tissue. The breakdown of elastic proteins in the lungs results in a loss of pulmonary resiliency and chronic lung hyperinflation. Because the walls between individual alveoli are broken down, the alveolar sacs that remain are larger.Lung diseases can be diagnosed and monitored using spirometry. A spirometer measures the flow rate and volume of inhaled and exhaled air. In one pulmonary test, a patient is asked to exhale forcibly and completely into a spirometer after maximum inhalation. This test is done to measure the patient's forced expiratory volume in one second (FEV1) and forced vital capacity (FVC), the total amount of air exhaled in a single breath.
 Figure 1 Forced expiratory spirogram of a healthy individual
Figure 1 Forced expiratory spirogram of a healthy individualAdapted from T. Barreiro, "An Approach to Interpreting Spirometry." American Family Physician. ©2004 AAFP.
Which of the following FEV1 spirograms would be expected in a patient with asthma?
A)

B)

C)

D)


Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
64
Passage
Obstructive respiratory illnesses are characterized by an increased difficulty exhaling much of the air in the lungs. Asthma and chronic obstructive pulmonary disease (COPD) are two of the most common obstructive respiratory illnesses. Asthma is due to allergen hypersensitivity and can be identified by temporary airway inflammation, which causes the bronchioles to narrow. The decreased airway diameter increases the resistance to airflow. Subsequently, air becomes "trapped" in the lungs, restricting the amount of air that can be moved in each breath.COPD refers to a spectrum of disorders and can be a result of emphysema, the irreversible destruction of pulmonary tissue. The breakdown of elastic proteins in the lungs results in a loss of pulmonary resiliency and chronic lung hyperinflation. Because the walls between individual alveoli are broken down, the alveolar sacs that remain are larger.Lung diseases can be diagnosed and monitored using spirometry. A spirometer measures the flow rate and volume of inhaled and exhaled air. In one pulmonary test, a patient is asked to exhale forcibly and completely into a spirometer after maximum inhalation. This test is done to measure the patient's forced expiratory volume in one second (FEV1) and forced vital capacity (FVC), the total amount of air exhaled in a single breath.
 Figure 1 Forced expiratory spirogram of a healthy individual
Figure 1 Forced expiratory spirogram of a healthy individual
Adapted from T. Barreiro, "An Approach to Interpreting Spirometry." American Family Physician. ©2004 AAFP.
During an asthma attack, temporary bronchoconstriction would have what effect on blood pH, and what would be the expected homeostatic response?
A)Respiratory acidosis and an increased respiratory rate
B)Respiratory acidosis and a decreased respiratory rate
C)Respiratory alkalosis and an increased respiratory rate
D)Respiratory alkalosis and a decreased respiratory rate
Obstructive respiratory illnesses are characterized by an increased difficulty exhaling much of the air in the lungs. Asthma and chronic obstructive pulmonary disease (COPD) are two of the most common obstructive respiratory illnesses. Asthma is due to allergen hypersensitivity and can be identified by temporary airway inflammation, which causes the bronchioles to narrow. The decreased airway diameter increases the resistance to airflow. Subsequently, air becomes "trapped" in the lungs, restricting the amount of air that can be moved in each breath.COPD refers to a spectrum of disorders and can be a result of emphysema, the irreversible destruction of pulmonary tissue. The breakdown of elastic proteins in the lungs results in a loss of pulmonary resiliency and chronic lung hyperinflation. Because the walls between individual alveoli are broken down, the alveolar sacs that remain are larger.Lung diseases can be diagnosed and monitored using spirometry. A spirometer measures the flow rate and volume of inhaled and exhaled air. In one pulmonary test, a patient is asked to exhale forcibly and completely into a spirometer after maximum inhalation. This test is done to measure the patient's forced expiratory volume in one second (FEV1) and forced vital capacity (FVC), the total amount of air exhaled in a single breath.
 Figure 1 Forced expiratory spirogram of a healthy individual
Figure 1 Forced expiratory spirogram of a healthy individualAdapted from T. Barreiro, "An Approach to Interpreting Spirometry." American Family Physician. ©2004 AAFP.
During an asthma attack, temporary bronchoconstriction would have what effect on blood pH, and what would be the expected homeostatic response?
A)Respiratory acidosis and an increased respiratory rate
B)Respiratory acidosis and a decreased respiratory rate
C)Respiratory alkalosis and an increased respiratory rate
D)Respiratory alkalosis and a decreased respiratory rate

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
65
Passage
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
 Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
 Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Adapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
During the interval between 30 and 60 minutes in Figure 1A, which of the following will be observed in WT mice?
A)Increased glucagon release.
B)Increased plasma amino acids.
C)Increased hepatic glycogen stores.
D)Increased hepatic gluconeogenesis.
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene. Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3). Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNAAdapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
During the interval between 30 and 60 minutes in Figure 1A, which of the following will be observed in WT mice?
A)Increased glucagon release.
B)Increased plasma amino acids.
C)Increased hepatic glycogen stores.
D)Increased hepatic gluconeogenesis.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
66
Passage
Mitochondria are thought to have been independent bacterial organisms that were engulfed and integrated into eukaryotic cells approximately two billion years ago. Most of the mitochondrial genome was lost or transferred into the large central genome of the eukaryotic nucleus, leaving only a residual genome within each mitochondrion.Because multiple mitochondria are found in the cell, mitochondrial DNA (mtDNA) mutations result in a condition known as heteroplasmy, or the intracellular mixture of wild-type mtDNA and mutant mtDNA. Just as the cellular ratio of wild-type to mutant mtDNA varies, the overall mtDNA content within a tissue or organ is also highly variable. For this reason, disease-causing mutant mtDNA manifests phenotypically only when the majority of mitochondria within a given tissue express the deleterious allele. In addition, the variability in mtDNA content makes it possible for individuals with the same mitochondrial mutation to display drastically different clinical symptoms.To investigate how heteroplasmy influences disease manifestation, researchers analyzed an A to G substitution at nucleotide 3243 in the tRNAleu gene of mtDNA. This mutation has been associated with hereditary hearing loss, diabetes mellitus, and a syndrome characterized by mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes.Family members who were confirmed to have the A3243G mutation were tested using an audiometry examination to assess hearing impairment, as shown in Table 1. The subjects carried no other genetic mutations known to cause hearing loss.Table 1 Audiometry Results From Family Members with the A3243G Mutation
 The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
 Figure 1 PAGE visualization of PCR products following ApaI digestion
Figure 1 PAGE visualization of PCR products following ApaI digestion
Adapted from Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc Biol Sci. 2012;279(1734):1865-72.
Which of the following would be true regarding the inheritance of the A3243G mutation? (Note: Assume affected individuals inherited the mutation.)
A)All daughters of an affected father are affected.
B)Only offspring of affected mothers are affected.
C)All affected males have asymptomatic carrier mothers.
D)Affected offspring must have two copies of the defective gene.
Mitochondria are thought to have been independent bacterial organisms that were engulfed and integrated into eukaryotic cells approximately two billion years ago. Most of the mitochondrial genome was lost or transferred into the large central genome of the eukaryotic nucleus, leaving only a residual genome within each mitochondrion.Because multiple mitochondria are found in the cell, mitochondrial DNA (mtDNA) mutations result in a condition known as heteroplasmy, or the intracellular mixture of wild-type mtDNA and mutant mtDNA. Just as the cellular ratio of wild-type to mutant mtDNA varies, the overall mtDNA content within a tissue or organ is also highly variable. For this reason, disease-causing mutant mtDNA manifests phenotypically only when the majority of mitochondria within a given tissue express the deleterious allele. In addition, the variability in mtDNA content makes it possible for individuals with the same mitochondrial mutation to display drastically different clinical symptoms.To investigate how heteroplasmy influences disease manifestation, researchers analyzed an A to G substitution at nucleotide 3243 in the tRNAleu gene of mtDNA. This mutation has been associated with hereditary hearing loss, diabetes mellitus, and a syndrome characterized by mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes.Family members who were confirmed to have the A3243G mutation were tested using an audiometry examination to assess hearing impairment, as shown in Table 1. The subjects carried no other genetic mutations known to cause hearing loss.Table 1 Audiometry Results From Family Members with the A3243G Mutation
 The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI. Figure 1 PAGE visualization of PCR products following ApaI digestion
Figure 1 PAGE visualization of PCR products following ApaI digestionAdapted from Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc Biol Sci. 2012;279(1734):1865-72.
Which of the following would be true regarding the inheritance of the A3243G mutation? (Note: Assume affected individuals inherited the mutation.)
A)All daughters of an affected father are affected.
B)Only offspring of affected mothers are affected.
C)All affected males have asymptomatic carrier mothers.
D)Affected offspring must have two copies of the defective gene.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
67
Passage
Obstructive respiratory illnesses are characterized by an increased difficulty exhaling much of the air in the lungs. Asthma and chronic obstructive pulmonary disease (COPD) are two of the most common obstructive respiratory illnesses. Asthma is due to allergen hypersensitivity and can be identified by temporary airway inflammation, which causes the bronchioles to narrow. The decreased airway diameter increases the resistance to airflow. Subsequently, air becomes "trapped" in the lungs, restricting the amount of air that can be moved in each breath.COPD refers to a spectrum of disorders and can be a result of emphysema, the irreversible destruction of pulmonary tissue. The breakdown of elastic proteins in the lungs results in a loss of pulmonary resiliency and chronic lung hyperinflation. Because the walls between individual alveoli are broken down, the alveolar sacs that remain are larger.Lung diseases can be diagnosed and monitored using spirometry. A spirometer measures the flow rate and volume of inhaled and exhaled air. In one pulmonary test, a patient is asked to exhale forcibly and completely into a spirometer after maximum inhalation. This test is done to measure the patient's forced expiratory volume in one second (FEV1) and forced vital capacity (FVC), the total amount of air exhaled in a single breath.
 Figure 1 Forced expiratory spirogram of a healthy individual
Figure 1 Forced expiratory spirogram of a healthy individual
Adapted from T. Barreiro, "An Approach to Interpreting Spirometry." American Family Physician. ©2004 AAFP.
The regulation of respiratory rate is normally most sensitive to:
A)PO2 in the blood.
B)PCO2 in the blood.
C)PO2 in the alveoli.
D)PCO2 in the alveoli.
Obstructive respiratory illnesses are characterized by an increased difficulty exhaling much of the air in the lungs. Asthma and chronic obstructive pulmonary disease (COPD) are two of the most common obstructive respiratory illnesses. Asthma is due to allergen hypersensitivity and can be identified by temporary airway inflammation, which causes the bronchioles to narrow. The decreased airway diameter increases the resistance to airflow. Subsequently, air becomes "trapped" in the lungs, restricting the amount of air that can be moved in each breath.COPD refers to a spectrum of disorders and can be a result of emphysema, the irreversible destruction of pulmonary tissue. The breakdown of elastic proteins in the lungs results in a loss of pulmonary resiliency and chronic lung hyperinflation. Because the walls between individual alveoli are broken down, the alveolar sacs that remain are larger.Lung diseases can be diagnosed and monitored using spirometry. A spirometer measures the flow rate and volume of inhaled and exhaled air. In one pulmonary test, a patient is asked to exhale forcibly and completely into a spirometer after maximum inhalation. This test is done to measure the patient's forced expiratory volume in one second (FEV1) and forced vital capacity (FVC), the total amount of air exhaled in a single breath.
 Figure 1 Forced expiratory spirogram of a healthy individual
Figure 1 Forced expiratory spirogram of a healthy individualAdapted from T. Barreiro, "An Approach to Interpreting Spirometry." American Family Physician. ©2004 AAFP.
The regulation of respiratory rate is normally most sensitive to:
A)PO2 in the blood.
B)PCO2 in the blood.
C)PO2 in the alveoli.
D)PCO2 in the alveoli.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
68
Passage
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
 Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
 Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Adapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
The researchers hypothesize that miR-26a overexpression can rescue obesity-induced insulin resistance. Which experimental result would NOT support their hypothesis?
A)Alb Tg mice fed a HFD had the same levels of random and fasting insulin as WT mice fed a SHD.
B)Alb Tg mice fed a HFD had the same levels of random and fasting glucose as WT mice fed a SHD.
C)WT mice fed a HFD had higher insulin sensitivity than Alb Tg mice fed a HFD.
D)WT mice fed a HFD had lower glucose tolerance than Alb Tg mice fed a HFD.
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene. Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3). Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNAAdapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
The researchers hypothesize that miR-26a overexpression can rescue obesity-induced insulin resistance. Which experimental result would NOT support their hypothesis?
A)Alb Tg mice fed a HFD had the same levels of random and fasting insulin as WT mice fed a SHD.
B)Alb Tg mice fed a HFD had the same levels of random and fasting glucose as WT mice fed a SHD.
C)WT mice fed a HFD had higher insulin sensitivity than Alb Tg mice fed a HFD.
D)WT mice fed a HFD had lower glucose tolerance than Alb Tg mice fed a HFD.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
69
Passage
Mitochondria are thought to have been independent bacterial organisms that were engulfed and integrated into eukaryotic cells approximately two billion years ago. Most of the mitochondrial genome was lost or transferred into the large central genome of the eukaryotic nucleus, leaving only a residual genome within each mitochondrion.Because multiple mitochondria are found in the cell, mitochondrial DNA (mtDNA) mutations result in a condition known as heteroplasmy, or the intracellular mixture of wild-type mtDNA and mutant mtDNA. Just as the cellular ratio of wild-type to mutant mtDNA varies, the overall mtDNA content within a tissue or organ is also highly variable. For this reason, disease-causing mutant mtDNA manifests phenotypically only when the majority of mitochondria within a given tissue express the deleterious allele. In addition, the variability in mtDNA content makes it possible for individuals with the same mitochondrial mutation to display drastically different clinical symptoms.To investigate how heteroplasmy influences disease manifestation, researchers analyzed an A to G substitution at nucleotide 3243 in the tRNAleu gene of mtDNA. This mutation has been associated with hereditary hearing loss, diabetes mellitus, and a syndrome characterized by mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes.Family members who were confirmed to have the A3243G mutation were tested using an audiometry examination to assess hearing impairment, as shown in Table 1. The subjects carried no other genetic mutations known to cause hearing loss.Table 1 Audiometry Results From Family Members with the A3243G Mutation
 The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
 Figure 1 PAGE visualization of PCR products following ApaI digestion
Figure 1 PAGE visualization of PCR products following ApaI digestion
Adapted from Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc Biol Sci. 2012;279(1734):1865-72.
The image below shows a population of unicellular organisms. In each organism, the shaded mitochondria express a deleterious mutant allele of a mitochondrial gene, and the nonshaded mitochondria express the wild-type allele of that same gene. Which of the following events most likely occurred between Generation 1 and Generation 2?
Which of the following events most likely occurred between Generation 1 and Generation 2?
A)Meiosis
B)Population bottleneck
C)Natural selection
D)New mutation
Mitochondria are thought to have been independent bacterial organisms that were engulfed and integrated into eukaryotic cells approximately two billion years ago. Most of the mitochondrial genome was lost or transferred into the large central genome of the eukaryotic nucleus, leaving only a residual genome within each mitochondrion.Because multiple mitochondria are found in the cell, mitochondrial DNA (mtDNA) mutations result in a condition known as heteroplasmy, or the intracellular mixture of wild-type mtDNA and mutant mtDNA. Just as the cellular ratio of wild-type to mutant mtDNA varies, the overall mtDNA content within a tissue or organ is also highly variable. For this reason, disease-causing mutant mtDNA manifests phenotypically only when the majority of mitochondria within a given tissue express the deleterious allele. In addition, the variability in mtDNA content makes it possible for individuals with the same mitochondrial mutation to display drastically different clinical symptoms.To investigate how heteroplasmy influences disease manifestation, researchers analyzed an A to G substitution at nucleotide 3243 in the tRNAleu gene of mtDNA. This mutation has been associated with hereditary hearing loss, diabetes mellitus, and a syndrome characterized by mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes.Family members who were confirmed to have the A3243G mutation were tested using an audiometry examination to assess hearing impairment, as shown in Table 1. The subjects carried no other genetic mutations known to cause hearing loss.Table 1 Audiometry Results From Family Members with the A3243G Mutation
 The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI. Figure 1 PAGE visualization of PCR products following ApaI digestion
Figure 1 PAGE visualization of PCR products following ApaI digestionAdapted from Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc Biol Sci. 2012;279(1734):1865-72.
The image below shows a population of unicellular organisms. In each organism, the shaded mitochondria express a deleterious mutant allele of a mitochondrial gene, and the nonshaded mitochondria express the wild-type allele of that same gene.
 Which of the following events most likely occurred between Generation 1 and Generation 2?
Which of the following events most likely occurred between Generation 1 and Generation 2?A)Meiosis
B)Population bottleneck
C)Natural selection
D)New mutation

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
70
Passage
Obstructive respiratory illnesses are characterized by an increased difficulty exhaling much of the air in the lungs. Asthma and chronic obstructive pulmonary disease (COPD) are two of the most common obstructive respiratory illnesses. Asthma is due to allergen hypersensitivity and can be identified by temporary airway inflammation, which causes the bronchioles to narrow. The decreased airway diameter increases the resistance to airflow. Subsequently, air becomes "trapped" in the lungs, restricting the amount of air that can be moved in each breath.COPD refers to a spectrum of disorders and can be a result of emphysema, the irreversible destruction of pulmonary tissue. The breakdown of elastic proteins in the lungs results in a loss of pulmonary resiliency and chronic lung hyperinflation. Because the walls between individual alveoli are broken down, the alveolar sacs that remain are larger.Lung diseases can be diagnosed and monitored using spirometry. A spirometer measures the flow rate and volume of inhaled and exhaled air. In one pulmonary test, a patient is asked to exhale forcibly and completely into a spirometer after maximum inhalation. This test is done to measure the patient's forced expiratory volume in one second (FEV1) and forced vital capacity (FVC), the total amount of air exhaled in a single breath.
 Figure 1 Forced expiratory spirogram of a healthy individual
Figure 1 Forced expiratory spirogram of a healthy individual
Adapted from T. Barreiro, "An Approach to Interpreting Spirometry." American Family Physician. ©2004 AAFP.
The inflation of the lungs in normal inspiration involves:Contraction of the diaphragmReduction of intrapleural pressureElevation of the rib cage
A)I and II only
B)I and III only
C)II and III only
D)I, II, and III
Obstructive respiratory illnesses are characterized by an increased difficulty exhaling much of the air in the lungs. Asthma and chronic obstructive pulmonary disease (COPD) are two of the most common obstructive respiratory illnesses. Asthma is due to allergen hypersensitivity and can be identified by temporary airway inflammation, which causes the bronchioles to narrow. The decreased airway diameter increases the resistance to airflow. Subsequently, air becomes "trapped" in the lungs, restricting the amount of air that can be moved in each breath.COPD refers to a spectrum of disorders and can be a result of emphysema, the irreversible destruction of pulmonary tissue. The breakdown of elastic proteins in the lungs results in a loss of pulmonary resiliency and chronic lung hyperinflation. Because the walls between individual alveoli are broken down, the alveolar sacs that remain are larger.Lung diseases can be diagnosed and monitored using spirometry. A spirometer measures the flow rate and volume of inhaled and exhaled air. In one pulmonary test, a patient is asked to exhale forcibly and completely into a spirometer after maximum inhalation. This test is done to measure the patient's forced expiratory volume in one second (FEV1) and forced vital capacity (FVC), the total amount of air exhaled in a single breath.
 Figure 1 Forced expiratory spirogram of a healthy individual
Figure 1 Forced expiratory spirogram of a healthy individualAdapted from T. Barreiro, "An Approach to Interpreting Spirometry." American Family Physician. ©2004 AAFP.
The inflation of the lungs in normal inspiration involves:Contraction of the diaphragmReduction of intrapleural pressureElevation of the rib cage
A)I and II only
B)I and III only
C)II and III only
D)I, II, and III

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
71
Passage
Eukaryotic organelles such as mitochondria and chloroplasts are believed to be relics of formerly free-living prokaryotes. The transition from a hypoxic (low O2) to an oxic atmosphere (21% O2) is said to have enabled primitive eukaryotic anaerobes to engulf ancient aerobic prokaryotes and consequently acquire the ability to produce energy through oxidative phosphorylation. This endosymbiotic theory of eukaryotic evolution also postulates that endosymbiosis resulted in larger eukaryotic genomes, which originated from the partial transfer of mitochondrial genes to the nuclear genome. On integration into the host genome, mitochondria-derived genes became indistinguishable from the original nuclear genes.Researchers have alternatively proposed that after a prolonged period of symbiosis, there is a possibility of gene transfer from eukaryotes to prokaryotes. This hypothesis was initially supported when copper/zinc (Cu/Zn) superoxide dismutase (SOD), a metalloprotein confined to the cytosol of eukaryotic cells, was found in Photobacterium leiognathi. The free-living bacterium P. leiognathi is also a known symbiont of ponyfish, a small fish species native to the Indian and Pacific Oceans.SODs are antioxidant enzymes that serve as the cell's first line of defense against reactive oxygen species (ROS). ROS produced by the electron transport chain damage proteins by oxidizing amino acid residues and metal ions on prosthetic groups, but can accumulate during times of biochemical and environmental stress. Superoxide (O2−) radicals, a form of ROS, are sequestered by SODs and converted into less toxic hydrogen peroxide (H2O2) and O2 gas.
Adapted from Bannister JV, Parker MW. The presence of a copper/zinc superoxide dismutase in the bacterium Photobacterium leiognathi: a likely case of gene transfer from eukaryotes to prokaryotes. Proc Natl Acad Sci USA. 1985;82(1):149-52.
Ponyfish cells containing P. leiognathi symbionts were exposed to a spindle fiber toxin that inhibits microtubule polymerization. Given this information, which of the following would most likely result as a consequence of toxin exposure?
A)P. leiognathi daughter cells with multiple copies of the Cu/Zn SOD gene
B)Ponyfish daughter cells containing the same copy number of the Cu/Zn SOD gene
C)Delayed separation of P. leiognathi cells during binary fission
D)Nondisjunction in somatic ponyfish cells undergoing nuclear division
Eukaryotic organelles such as mitochondria and chloroplasts are believed to be relics of formerly free-living prokaryotes. The transition from a hypoxic (low O2) to an oxic atmosphere (21% O2) is said to have enabled primitive eukaryotic anaerobes to engulf ancient aerobic prokaryotes and consequently acquire the ability to produce energy through oxidative phosphorylation. This endosymbiotic theory of eukaryotic evolution also postulates that endosymbiosis resulted in larger eukaryotic genomes, which originated from the partial transfer of mitochondrial genes to the nuclear genome. On integration into the host genome, mitochondria-derived genes became indistinguishable from the original nuclear genes.Researchers have alternatively proposed that after a prolonged period of symbiosis, there is a possibility of gene transfer from eukaryotes to prokaryotes. This hypothesis was initially supported when copper/zinc (Cu/Zn) superoxide dismutase (SOD), a metalloprotein confined to the cytosol of eukaryotic cells, was found in Photobacterium leiognathi. The free-living bacterium P. leiognathi is also a known symbiont of ponyfish, a small fish species native to the Indian and Pacific Oceans.SODs are antioxidant enzymes that serve as the cell's first line of defense against reactive oxygen species (ROS). ROS produced by the electron transport chain damage proteins by oxidizing amino acid residues and metal ions on prosthetic groups, but can accumulate during times of biochemical and environmental stress. Superoxide (O2−) radicals, a form of ROS, are sequestered by SODs and converted into less toxic hydrogen peroxide (H2O2) and O2 gas.
Adapted from Bannister JV, Parker MW. The presence of a copper/zinc superoxide dismutase in the bacterium Photobacterium leiognathi: a likely case of gene transfer from eukaryotes to prokaryotes. Proc Natl Acad Sci USA. 1985;82(1):149-52.
Ponyfish cells containing P. leiognathi symbionts were exposed to a spindle fiber toxin that inhibits microtubule polymerization. Given this information, which of the following would most likely result as a consequence of toxin exposure?
A)P. leiognathi daughter cells with multiple copies of the Cu/Zn SOD gene
B)Ponyfish daughter cells containing the same copy number of the Cu/Zn SOD gene
C)Delayed separation of P. leiognathi cells during binary fission
D)Nondisjunction in somatic ponyfish cells undergoing nuclear division

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
72
Passage
Mitochondria are thought to have been independent bacterial organisms that were engulfed and integrated into eukaryotic cells approximately two billion years ago. Most of the mitochondrial genome was lost or transferred into the large central genome of the eukaryotic nucleus, leaving only a residual genome within each mitochondrion.Because multiple mitochondria are found in the cell, mitochondrial DNA (mtDNA) mutations result in a condition known as heteroplasmy, or the intracellular mixture of wild-type mtDNA and mutant mtDNA. Just as the cellular ratio of wild-type to mutant mtDNA varies, the overall mtDNA content within a tissue or organ is also highly variable. For this reason, disease-causing mutant mtDNA manifests phenotypically only when the majority of mitochondria within a given tissue express the deleterious allele. In addition, the variability in mtDNA content makes it possible for individuals with the same mitochondrial mutation to display drastically different clinical symptoms.To investigate how heteroplasmy influences disease manifestation, researchers analyzed an A to G substitution at nucleotide 3243 in the tRNAleu gene of mtDNA. This mutation has been associated with hereditary hearing loss, diabetes mellitus, and a syndrome characterized by mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes.Family members who were confirmed to have the A3243G mutation were tested using an audiometry examination to assess hearing impairment, as shown in Table 1. The subjects carried no other genetic mutations known to cause hearing loss.Table 1 Audiometry Results From Family Members with the A3243G Mutation
 The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
 Figure 1 PAGE visualization of PCR products following ApaI digestion
Figure 1 PAGE visualization of PCR products following ApaI digestion
Adapted from Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc Biol Sci. 2012;279(1734):1865-72.
Based on the passage, which of the following reagents was most likely used during PCR?
A)Free ribonucleoside triphosphates
B)RNA polymerase
C)A template sequence containing deoxyribonucleotides
D)Primer pairs with minimal GC content
Mitochondria are thought to have been independent bacterial organisms that were engulfed and integrated into eukaryotic cells approximately two billion years ago. Most of the mitochondrial genome was lost or transferred into the large central genome of the eukaryotic nucleus, leaving only a residual genome within each mitochondrion.Because multiple mitochondria are found in the cell, mitochondrial DNA (mtDNA) mutations result in a condition known as heteroplasmy, or the intracellular mixture of wild-type mtDNA and mutant mtDNA. Just as the cellular ratio of wild-type to mutant mtDNA varies, the overall mtDNA content within a tissue or organ is also highly variable. For this reason, disease-causing mutant mtDNA manifests phenotypically only when the majority of mitochondria within a given tissue express the deleterious allele. In addition, the variability in mtDNA content makes it possible for individuals with the same mitochondrial mutation to display drastically different clinical symptoms.To investigate how heteroplasmy influences disease manifestation, researchers analyzed an A to G substitution at nucleotide 3243 in the tRNAleu gene of mtDNA. This mutation has been associated with hereditary hearing loss, diabetes mellitus, and a syndrome characterized by mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes.Family members who were confirmed to have the A3243G mutation were tested using an audiometry examination to assess hearing impairment, as shown in Table 1. The subjects carried no other genetic mutations known to cause hearing loss.Table 1 Audiometry Results From Family Members with the A3243G Mutation
 The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI. Figure 1 PAGE visualization of PCR products following ApaI digestion
Figure 1 PAGE visualization of PCR products following ApaI digestionAdapted from Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc Biol Sci. 2012;279(1734):1865-72.
Based on the passage, which of the following reagents was most likely used during PCR?
A)Free ribonucleoside triphosphates
B)RNA polymerase
C)A template sequence containing deoxyribonucleotides
D)Primer pairs with minimal GC content

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
73
Passage
Eukaryotic organelles such as mitochondria and chloroplasts are believed to be relics of formerly free-living prokaryotes. The transition from a hypoxic (low O2) to an oxic atmosphere (21% O2) is said to have enabled primitive eukaryotic anaerobes to engulf ancient aerobic prokaryotes and consequently acquire the ability to produce energy through oxidative phosphorylation. This endosymbiotic theory of eukaryotic evolution also postulates that endosymbiosis resulted in larger eukaryotic genomes, which originated from the partial transfer of mitochondrial genes to the nuclear genome. On integration into the host genome, mitochondria-derived genes became indistinguishable from the original nuclear genes.Researchers have alternatively proposed that after a prolonged period of symbiosis, there is a possibility of gene transfer from eukaryotes to prokaryotes. This hypothesis was initially supported when copper/zinc (Cu/Zn) superoxide dismutase (SOD), a metalloprotein confined to the cytosol of eukaryotic cells, was found in Photobacterium leiognathi. The free-living bacterium P. leiognathi is also a known symbiont of ponyfish, a small fish species native to the Indian and Pacific Oceans.SODs are antioxidant enzymes that serve as the cell's first line of defense against reactive oxygen species (ROS). ROS produced by the electron transport chain damage proteins by oxidizing amino acid residues and metal ions on prosthetic groups, but can accumulate during times of biochemical and environmental stress. Superoxide (O2−) radicals, a form of ROS, are sequestered by SODs and converted into less toxic hydrogen peroxide (H2O2) and O2 gas.
Adapted from Bannister JV, Parker MW. The presence of a copper/zinc superoxide dismutase in the bacterium Photobacterium leiognathi: a likely case of gene transfer from eukaryotes to prokaryotes. Proc Natl Acad Sci USA. 1985;82(1):149-52.
Based on the passage, which of the following best explains how mitochondrial genes differed from eukaryotic genes prior to gene transfer? Mitochondrial genes were:
A)interspersed with noncoding sequences.
B)located on chromosomes without telomeres.
C)present on a single-stranded DNA genome.
D)associated with histones.
Eukaryotic organelles such as mitochondria and chloroplasts are believed to be relics of formerly free-living prokaryotes. The transition from a hypoxic (low O2) to an oxic atmosphere (21% O2) is said to have enabled primitive eukaryotic anaerobes to engulf ancient aerobic prokaryotes and consequently acquire the ability to produce energy through oxidative phosphorylation. This endosymbiotic theory of eukaryotic evolution also postulates that endosymbiosis resulted in larger eukaryotic genomes, which originated from the partial transfer of mitochondrial genes to the nuclear genome. On integration into the host genome, mitochondria-derived genes became indistinguishable from the original nuclear genes.Researchers have alternatively proposed that after a prolonged period of symbiosis, there is a possibility of gene transfer from eukaryotes to prokaryotes. This hypothesis was initially supported when copper/zinc (Cu/Zn) superoxide dismutase (SOD), a metalloprotein confined to the cytosol of eukaryotic cells, was found in Photobacterium leiognathi. The free-living bacterium P. leiognathi is also a known symbiont of ponyfish, a small fish species native to the Indian and Pacific Oceans.SODs are antioxidant enzymes that serve as the cell's first line of defense against reactive oxygen species (ROS). ROS produced by the electron transport chain damage proteins by oxidizing amino acid residues and metal ions on prosthetic groups, but can accumulate during times of biochemical and environmental stress. Superoxide (O2−) radicals, a form of ROS, are sequestered by SODs and converted into less toxic hydrogen peroxide (H2O2) and O2 gas.
Adapted from Bannister JV, Parker MW. The presence of a copper/zinc superoxide dismutase in the bacterium Photobacterium leiognathi: a likely case of gene transfer from eukaryotes to prokaryotes. Proc Natl Acad Sci USA. 1985;82(1):149-52.
Based on the passage, which of the following best explains how mitochondrial genes differed from eukaryotic genes prior to gene transfer? Mitochondrial genes were:
A)interspersed with noncoding sequences.
B)located on chromosomes without telomeres.
C)present on a single-stranded DNA genome.
D)associated with histones.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
74
Passage
Obstructive respiratory illnesses are characterized by an increased difficulty exhaling much of the air in the lungs. Asthma and chronic obstructive pulmonary disease (COPD) are two of the most common obstructive respiratory illnesses. Asthma is due to allergen hypersensitivity and can be identified by temporary airway inflammation, which causes the bronchioles to narrow. The decreased airway diameter increases the resistance to airflow. Subsequently, air becomes "trapped" in the lungs, restricting the amount of air that can be moved in each breath.COPD refers to a spectrum of disorders and can be a result of emphysema, the irreversible destruction of pulmonary tissue. The breakdown of elastic proteins in the lungs results in a loss of pulmonary resiliency and chronic lung hyperinflation. Because the walls between individual alveoli are broken down, the alveolar sacs that remain are larger.Lung diseases can be diagnosed and monitored using spirometry. A spirometer measures the flow rate and volume of inhaled and exhaled air. In one pulmonary test, a patient is asked to exhale forcibly and completely into a spirometer after maximum inhalation. This test is done to measure the patient's forced expiratory volume in one second (FEV1) and forced vital capacity (FVC), the total amount of air exhaled in a single breath.
 Figure 1 Forced expiratory spirogram of a healthy individual
Figure 1 Forced expiratory spirogram of a healthy individual
Adapted from T. Barreiro, "An Approach to Interpreting Spirometry." American Family Physician. ©2004 AAFP.
Primary ciliary dyskinesia (PCD), a rare disease that impairs the normal function of cilia, is often misdiagnosed as atypical asthma because wheezing is observed in both illnesses. Unlike asthma, PCD would cause the inability to:
A)balance respiratory thermoregulation.
B)cough or sneeze.
C)remove inhaled particulates.
D)speak in lower pitches.
Obstructive respiratory illnesses are characterized by an increased difficulty exhaling much of the air in the lungs. Asthma and chronic obstructive pulmonary disease (COPD) are two of the most common obstructive respiratory illnesses. Asthma is due to allergen hypersensitivity and can be identified by temporary airway inflammation, which causes the bronchioles to narrow. The decreased airway diameter increases the resistance to airflow. Subsequently, air becomes "trapped" in the lungs, restricting the amount of air that can be moved in each breath.COPD refers to a spectrum of disorders and can be a result of emphysema, the irreversible destruction of pulmonary tissue. The breakdown of elastic proteins in the lungs results in a loss of pulmonary resiliency and chronic lung hyperinflation. Because the walls between individual alveoli are broken down, the alveolar sacs that remain are larger.Lung diseases can be diagnosed and monitored using spirometry. A spirometer measures the flow rate and volume of inhaled and exhaled air. In one pulmonary test, a patient is asked to exhale forcibly and completely into a spirometer after maximum inhalation. This test is done to measure the patient's forced expiratory volume in one second (FEV1) and forced vital capacity (FVC), the total amount of air exhaled in a single breath.
 Figure 1 Forced expiratory spirogram of a healthy individual
Figure 1 Forced expiratory spirogram of a healthy individualAdapted from T. Barreiro, "An Approach to Interpreting Spirometry." American Family Physician. ©2004 AAFP.
Primary ciliary dyskinesia (PCD), a rare disease that impairs the normal function of cilia, is often misdiagnosed as atypical asthma because wheezing is observed in both illnesses. Unlike asthma, PCD would cause the inability to:
A)balance respiratory thermoregulation.
B)cough or sneeze.
C)remove inhaled particulates.
D)speak in lower pitches.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
75
Passage
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
 Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
 Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Adapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
One of the study subjects with a BMI of 56 kg/m2 (Figure 2) is discovered to have a similar pathology to the ob/ob mouse. Like ob/ob mice, affected individuals exhibit normal weight at birth but become severely obese afterward and have undetectable levels of leptin in the serum. What is the best explanation for the absence of serum leptin in this condition?
A)Leptin cannot bind its receptor in the hypothalamus.
B)Leptin cannot bind its receptor in the thyroid gland.
C)Leptin cannot be secreted from gastric cells.
D)Leptin cannot be secreted from adipocytes.
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene. Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3). Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNAAdapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
One of the study subjects with a BMI of 56 kg/m2 (Figure 2) is discovered to have a similar pathology to the ob/ob mouse. Like ob/ob mice, affected individuals exhibit normal weight at birth but become severely obese afterward and have undetectable levels of leptin in the serum. What is the best explanation for the absence of serum leptin in this condition?
A)Leptin cannot bind its receptor in the hypothalamus.
B)Leptin cannot bind its receptor in the thyroid gland.
C)Leptin cannot be secreted from gastric cells.
D)Leptin cannot be secreted from adipocytes.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
76
Passage
Eukaryotic organelles such as mitochondria and chloroplasts are believed to be relics of formerly free-living prokaryotes. The transition from a hypoxic (low O2) to an oxic atmosphere (21% O2) is said to have enabled primitive eukaryotic anaerobes to engulf ancient aerobic prokaryotes and consequently acquire the ability to produce energy through oxidative phosphorylation. This endosymbiotic theory of eukaryotic evolution also postulates that endosymbiosis resulted in larger eukaryotic genomes, which originated from the partial transfer of mitochondrial genes to the nuclear genome. On integration into the host genome, mitochondria-derived genes became indistinguishable from the original nuclear genes.Researchers have alternatively proposed that after a prolonged period of symbiosis, there is a possibility of gene transfer from eukaryotes to prokaryotes. This hypothesis was initially supported when copper/zinc (Cu/Zn) superoxide dismutase (SOD), a metalloprotein confined to the cytosol of eukaryotic cells, was found in Photobacterium leiognathi. The free-living bacterium P. leiognathi is also a known symbiont of ponyfish, a small fish species native to the Indian and Pacific Oceans.SODs are antioxidant enzymes that serve as the cell's first line of defense against reactive oxygen species (ROS). ROS produced by the electron transport chain damage proteins by oxidizing amino acid residues and metal ions on prosthetic groups, but can accumulate during times of biochemical and environmental stress. Superoxide (O2−) radicals, a form of ROS, are sequestered by SODs and converted into less toxic hydrogen peroxide (H2O2) and O2 gas.
Adapted from Bannister JV, Parker MW. The presence of a copper/zinc superoxide dismutase in the bacterium Photobacterium leiognathi: a likely case of gene transfer from eukaryotes to prokaryotes. Proc Natl Acad Sci USA. 1985;82(1):149-52.
SOD mRNA in P. leiognathi encodes a signal sequence that directs the transport of the SOD protein for secretion. This signal sequence will direct SOD proteins to which structure in P. leiognathi?
A)Smooth endoplasmic reticulum
B)Plasma membrane
C)Mitochondrial outer membrane
D)Golgi body
Eukaryotic organelles such as mitochondria and chloroplasts are believed to be relics of formerly free-living prokaryotes. The transition from a hypoxic (low O2) to an oxic atmosphere (21% O2) is said to have enabled primitive eukaryotic anaerobes to engulf ancient aerobic prokaryotes and consequently acquire the ability to produce energy through oxidative phosphorylation. This endosymbiotic theory of eukaryotic evolution also postulates that endosymbiosis resulted in larger eukaryotic genomes, which originated from the partial transfer of mitochondrial genes to the nuclear genome. On integration into the host genome, mitochondria-derived genes became indistinguishable from the original nuclear genes.Researchers have alternatively proposed that after a prolonged period of symbiosis, there is a possibility of gene transfer from eukaryotes to prokaryotes. This hypothesis was initially supported when copper/zinc (Cu/Zn) superoxide dismutase (SOD), a metalloprotein confined to the cytosol of eukaryotic cells, was found in Photobacterium leiognathi. The free-living bacterium P. leiognathi is also a known symbiont of ponyfish, a small fish species native to the Indian and Pacific Oceans.SODs are antioxidant enzymes that serve as the cell's first line of defense against reactive oxygen species (ROS). ROS produced by the electron transport chain damage proteins by oxidizing amino acid residues and metal ions on prosthetic groups, but can accumulate during times of biochemical and environmental stress. Superoxide (O2−) radicals, a form of ROS, are sequestered by SODs and converted into less toxic hydrogen peroxide (H2O2) and O2 gas.
Adapted from Bannister JV, Parker MW. The presence of a copper/zinc superoxide dismutase in the bacterium Photobacterium leiognathi: a likely case of gene transfer from eukaryotes to prokaryotes. Proc Natl Acad Sci USA. 1985;82(1):149-52.
SOD mRNA in P. leiognathi encodes a signal sequence that directs the transport of the SOD protein for secretion. This signal sequence will direct SOD proteins to which structure in P. leiognathi?
A)Smooth endoplasmic reticulum
B)Plasma membrane
C)Mitochondrial outer membrane
D)Golgi body

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
77
Passage
Mitochondria are thought to have been independent bacterial organisms that were engulfed and integrated into eukaryotic cells approximately two billion years ago. Most of the mitochondrial genome was lost or transferred into the large central genome of the eukaryotic nucleus, leaving only a residual genome within each mitochondrion.Because multiple mitochondria are found in the cell, mitochondrial DNA (mtDNA) mutations result in a condition known as heteroplasmy, or the intracellular mixture of wild-type mtDNA and mutant mtDNA. Just as the cellular ratio of wild-type to mutant mtDNA varies, the overall mtDNA content within a tissue or organ is also highly variable. For this reason, disease-causing mutant mtDNA manifests phenotypically only when the majority of mitochondria within a given tissue express the deleterious allele. In addition, the variability in mtDNA content makes it possible for individuals with the same mitochondrial mutation to display drastically different clinical symptoms.To investigate how heteroplasmy influences disease manifestation, researchers analyzed an A to G substitution at nucleotide 3243 in the tRNAleu gene of mtDNA. This mutation has been associated with hereditary hearing loss, diabetes mellitus, and a syndrome characterized by mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes.Family members who were confirmed to have the A3243G mutation were tested using an audiometry examination to assess hearing impairment, as shown in Table 1. The subjects carried no other genetic mutations known to cause hearing loss.Table 1 Audiometry Results From Family Members with the A3243G Mutation
 The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
 Figure 1 PAGE visualization of PCR products following ApaI digestion
Figure 1 PAGE visualization of PCR products following ApaI digestion
Adapted from Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc Biol Sci. 2012;279(1734):1865-72.
Which of the following cell types would be LEAST sensitive to mitochondrial dysfunction due to mtDNA mutation?
A)Hepatocytes
B)Neurons
C)Myocytes
D)Erythrocytes
Mitochondria are thought to have been independent bacterial organisms that were engulfed and integrated into eukaryotic cells approximately two billion years ago. Most of the mitochondrial genome was lost or transferred into the large central genome of the eukaryotic nucleus, leaving only a residual genome within each mitochondrion.Because multiple mitochondria are found in the cell, mitochondrial DNA (mtDNA) mutations result in a condition known as heteroplasmy, or the intracellular mixture of wild-type mtDNA and mutant mtDNA. Just as the cellular ratio of wild-type to mutant mtDNA varies, the overall mtDNA content within a tissue or organ is also highly variable. For this reason, disease-causing mutant mtDNA manifests phenotypically only when the majority of mitochondria within a given tissue express the deleterious allele. In addition, the variability in mtDNA content makes it possible for individuals with the same mitochondrial mutation to display drastically different clinical symptoms.To investigate how heteroplasmy influences disease manifestation, researchers analyzed an A to G substitution at nucleotide 3243 in the tRNAleu gene of mtDNA. This mutation has been associated with hereditary hearing loss, diabetes mellitus, and a syndrome characterized by mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes.Family members who were confirmed to have the A3243G mutation were tested using an audiometry examination to assess hearing impairment, as shown in Table 1. The subjects carried no other genetic mutations known to cause hearing loss.Table 1 Audiometry Results From Family Members with the A3243G Mutation
 The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI. Figure 1 PAGE visualization of PCR products following ApaI digestion
Figure 1 PAGE visualization of PCR products following ApaI digestionAdapted from Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc Biol Sci. 2012;279(1734):1865-72.
Which of the following cell types would be LEAST sensitive to mitochondrial dysfunction due to mtDNA mutation?
A)Hepatocytes
B)Neurons
C)Myocytes
D)Erythrocytes

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
78
Passage
Eukaryotic organelles such as mitochondria and chloroplasts are believed to be relics of formerly free-living prokaryotes. The transition from a hypoxic (low O2) to an oxic atmosphere (21% O2) is said to have enabled primitive eukaryotic anaerobes to engulf ancient aerobic prokaryotes and consequently acquire the ability to produce energy through oxidative phosphorylation. This endosymbiotic theory of eukaryotic evolution also postulates that endosymbiosis resulted in larger eukaryotic genomes, which originated from the partial transfer of mitochondrial genes to the nuclear genome. On integration into the host genome, mitochondria-derived genes became indistinguishable from the original nuclear genes.Researchers have alternatively proposed that after a prolonged period of symbiosis, there is a possibility of gene transfer from eukaryotes to prokaryotes. This hypothesis was initially supported when copper/zinc (Cu/Zn) superoxide dismutase (SOD), a metalloprotein confined to the cytosol of eukaryotic cells, was found in Photobacterium leiognathi. The free-living bacterium P. leiognathi is also a known symbiont of ponyfish, a small fish species native to the Indian and Pacific Oceans.SODs are antioxidant enzymes that serve as the cell's first line of defense against reactive oxygen species (ROS). ROS produced by the electron transport chain damage proteins by oxidizing amino acid residues and metal ions on prosthetic groups, but can accumulate during times of biochemical and environmental stress. Superoxide (O2−) radicals, a form of ROS, are sequestered by SODs and converted into less toxic hydrogen peroxide (H2O2) and O2 gas.
Adapted from Bannister JV, Parker MW. The presence of a copper/zinc superoxide dismutase in the bacterium Photobacterium leiognathi: a likely case of gene transfer from eukaryotes to prokaryotes. Proc Natl Acad Sci USA. 1985;82(1):149-52.
Which observation would NOT support the hypothesis of gene transfer from eukaryotes to prokaryotes described in the passage?
A)Discovering no difference between the gene sequences of the P. leiognathi and ponyfish SODs
B)Discovering P. leiognathi and ponyfish SODs have a similar amino acid sequence in their noncatalytic domains
C)Discovering Cu/Zn SODs in other free-living bacterial species with no known eukaryotic symbiotic hosts
D)Discovering Cu/Zn SODs in other free-living bacterial species with known eukaryotic symbiotic hosts
Eukaryotic organelles such as mitochondria and chloroplasts are believed to be relics of formerly free-living prokaryotes. The transition from a hypoxic (low O2) to an oxic atmosphere (21% O2) is said to have enabled primitive eukaryotic anaerobes to engulf ancient aerobic prokaryotes and consequently acquire the ability to produce energy through oxidative phosphorylation. This endosymbiotic theory of eukaryotic evolution also postulates that endosymbiosis resulted in larger eukaryotic genomes, which originated from the partial transfer of mitochondrial genes to the nuclear genome. On integration into the host genome, mitochondria-derived genes became indistinguishable from the original nuclear genes.Researchers have alternatively proposed that after a prolonged period of symbiosis, there is a possibility of gene transfer from eukaryotes to prokaryotes. This hypothesis was initially supported when copper/zinc (Cu/Zn) superoxide dismutase (SOD), a metalloprotein confined to the cytosol of eukaryotic cells, was found in Photobacterium leiognathi. The free-living bacterium P. leiognathi is also a known symbiont of ponyfish, a small fish species native to the Indian and Pacific Oceans.SODs are antioxidant enzymes that serve as the cell's first line of defense against reactive oxygen species (ROS). ROS produced by the electron transport chain damage proteins by oxidizing amino acid residues and metal ions on prosthetic groups, but can accumulate during times of biochemical and environmental stress. Superoxide (O2−) radicals, a form of ROS, are sequestered by SODs and converted into less toxic hydrogen peroxide (H2O2) and O2 gas.
Adapted from Bannister JV, Parker MW. The presence of a copper/zinc superoxide dismutase in the bacterium Photobacterium leiognathi: a likely case of gene transfer from eukaryotes to prokaryotes. Proc Natl Acad Sci USA. 1985;82(1):149-52.
Which observation would NOT support the hypothesis of gene transfer from eukaryotes to prokaryotes described in the passage?
A)Discovering no difference between the gene sequences of the P. leiognathi and ponyfish SODs
B)Discovering P. leiognathi and ponyfish SODs have a similar amino acid sequence in their noncatalytic domains
C)Discovering Cu/Zn SODs in other free-living bacterial species with no known eukaryotic symbiotic hosts
D)Discovering Cu/Zn SODs in other free-living bacterial species with known eukaryotic symbiotic hosts

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
79
Passage
Mitochondria are thought to have been independent bacterial organisms that were engulfed and integrated into eukaryotic cells approximately two billion years ago. Most of the mitochondrial genome was lost or transferred into the large central genome of the eukaryotic nucleus, leaving only a residual genome within each mitochondrion.Because multiple mitochondria are found in the cell, mitochondrial DNA (mtDNA) mutations result in a condition known as heteroplasmy, or the intracellular mixture of wild-type mtDNA and mutant mtDNA. Just as the cellular ratio of wild-type to mutant mtDNA varies, the overall mtDNA content within a tissue or organ is also highly variable. For this reason, disease-causing mutant mtDNA manifests phenotypically only when the majority of mitochondria within a given tissue express the deleterious allele. In addition, the variability in mtDNA content makes it possible for individuals with the same mitochondrial mutation to display drastically different clinical symptoms.To investigate how heteroplasmy influences disease manifestation, researchers analyzed an A to G substitution at nucleotide 3243 in the tRNAleu gene of mtDNA. This mutation has been associated with hereditary hearing loss, diabetes mellitus, and a syndrome characterized by mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes.Family members who were confirmed to have the A3243G mutation were tested using an audiometry examination to assess hearing impairment, as shown in Table 1. The subjects carried no other genetic mutations known to cause hearing loss.Table 1 Audiometry Results From Family Members with the A3243G Mutation
 The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
 Figure 1 PAGE visualization of PCR products following ApaI digestion
Figure 1 PAGE visualization of PCR products following ApaI digestion
Adapted from Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc Biol Sci. 2012;279(1734):1865-72.
Based on the information given in Table 1, the results of this examination may be explained by all of the following EXCEPT:
A)moderate hearing loss is influenced by autosomal recessivity at the 3243 locus.
B)severe hearing loss is influenced by a single nucleotide mutation.
C)the mutant tRNAleu gene causes variable expressivity of the phenotype.
D)the mutant tRNAleu gene exhibits incomplete penetrance.
Mitochondria are thought to have been independent bacterial organisms that were engulfed and integrated into eukaryotic cells approximately two billion years ago. Most of the mitochondrial genome was lost or transferred into the large central genome of the eukaryotic nucleus, leaving only a residual genome within each mitochondrion.Because multiple mitochondria are found in the cell, mitochondrial DNA (mtDNA) mutations result in a condition known as heteroplasmy, or the intracellular mixture of wild-type mtDNA and mutant mtDNA. Just as the cellular ratio of wild-type to mutant mtDNA varies, the overall mtDNA content within a tissue or organ is also highly variable. For this reason, disease-causing mutant mtDNA manifests phenotypically only when the majority of mitochondria within a given tissue express the deleterious allele. In addition, the variability in mtDNA content makes it possible for individuals with the same mitochondrial mutation to display drastically different clinical symptoms.To investigate how heteroplasmy influences disease manifestation, researchers analyzed an A to G substitution at nucleotide 3243 in the tRNAleu gene of mtDNA. This mutation has been associated with hereditary hearing loss, diabetes mellitus, and a syndrome characterized by mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes.Family members who were confirmed to have the A3243G mutation were tested using an audiometry examination to assess hearing impairment, as shown in Table 1. The subjects carried no other genetic mutations known to cause hearing loss.Table 1 Audiometry Results From Family Members with the A3243G Mutation
 The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI.
The blood samples of these subjects were collected, and the 3243 region of the tRNAleu gene was amplified via standard PCR. The PCR products were subsequently treated with the restriction enzyme ApaI and visualized on a PAGE gel. The normal undigested PCR product is 161 bp in length, but the PCR product amplified from the mutant A3243G is digested into two fragments by ApaI. Figure 1 PAGE visualization of PCR products following ApaI digestion
Figure 1 PAGE visualization of PCR products following ApaI digestionAdapted from Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc Biol Sci. 2012;279(1734):1865-72.
Based on the information given in Table 1, the results of this examination may be explained by all of the following EXCEPT:
A)moderate hearing loss is influenced by autosomal recessivity at the 3243 locus.
B)severe hearing loss is influenced by a single nucleotide mutation.
C)the mutant tRNAleu gene causes variable expressivity of the phenotype.
D)the mutant tRNAleu gene exhibits incomplete penetrance.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck
80
Passage
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
 Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
 Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Adapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
Recently fed ob/ob mice show deficient binding of glucagon to its G protein-coupled receptor, which most likely leads to:
A)decreased cAMP to ATP conversion.
B)upregulated adenylate cyclase activity.
C)increased exchange of GDP for GTP.
D)reduced protein kinase A activity.
Alzheimer disease (AD) is a progressive condition affecting memory and cognition that can be pathologically characterized by the presence of neurofibrillary tangles (NFTs). NFTs, aggregates of hyperphosphorylated tau proteins that have dissociated from microtubules, are believed to contribute to neuronal degradation and subsequent AD symptoms. In general, ribosomes initiate translation of tau mRNA by binding the 7-methylguanosine (m7Gpp) cap present at the end of the 5′ untranslated region (UTR) of the mRNA molecule. However, researchers believe that tau protein translation initiation can occur without recognition of the m7Gpp cap. This alternate mechanism, referred to as cap-independent translation, indicates that ribosomes are recruited by internal ribosome entry sites (IRES) generally located in the 5′ UTR of the mRNA sequence.To determine the initiation mechanism of tau mRNA translation, neuroblastoma cells of AD mice were transfected with two different DNA constructs (Figure 1). Construct 1 contained the 5′ UTR of tau mRNA isoforms (v1 and v2) extracted from cells displaying NFTs. Construct 2 included the 5′ UTR of the β-globulin gene, which normally codes for a plasma protein. The luciferase reporter genes, Renillla and Photinus, were also included in the two constructs.
/_
/_
 Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene.
Figure 1 DNA constructs containing β-globulin or tau gene used to transfect neuroblastoma cellsLuciferase gene expression from the constructs was monitored in the absence and presence of a synthetic m7Gpp cap analog (Figure 2). Translation of Renillla luciferase mRNA depended on ribosomal binding to the m7Gpp cap of the mRNA, but translation of Photinus luciferase required ribosome recruitment by an IRES in the 5′ UTR of the adjacent gene. Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3).
Figure 2 Luciferase activity in neuroblastoma cells exposed to m7Gpp cap analogIn a second study, researchers used siRNA to target initiation factor eIF4E, a peptide translated in a cap-dependent manner that facilitates the binding of ribosomes to the m7Gpp cap. Expression of factor eIF4E, elongation factor eEF2K, and tau v2 protein was assessed in siRNA-treated cells (Figure 3). Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNA
Figure 3 Western blot of eIF4E, eEF2K, and tau v2 extracted from non-AD cells treated with eIF4E siRNAAdapted from Veo BL, Krushel LA. Translation initiation of the human tau mRNA through an internal ribosomal entry site. J Alzheimers Dis. 2009; 16(2):271-5.
Recently fed ob/ob mice show deficient binding of glucagon to its G protein-coupled receptor, which most likely leads to:
A)decreased cAMP to ATP conversion.
B)upregulated adenylate cyclase activity.
C)increased exchange of GDP for GTP.
D)reduced protein kinase A activity.

Unlock Deck
Unlock for access to all 319 flashcards in this deck.
Unlock Deck
k this deck



