Deck 4: Antigen Recognition by B-Cell and T-Cell Receptors
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/28
Play
Full screen (f)
Deck 4: Antigen Recognition by B-Cell and T-Cell Receptors
1
The innate immune response together with antibodies are generally not effective at clearing infections established by pathogens that replicate inside host cells. The evolution of T cells has provided a means for the immune response to 'see' intracellular infections based on the ability of T cells to:
A) Secrete cytokines that diffuse into the infected tissue
B) Activate type I interferon production by macrophages and dendritic cells
C) Activate macrophages to induce inflammation
D) Recognize pathogen-derived peptides on host MHC surface molecules
E) Express cytoplasmic sensors for detecting pathogen-derived nucleic acids
A) Secrete cytokines that diffuse into the infected tissue
B) Activate type I interferon production by macrophages and dendritic cells
C) Activate macrophages to induce inflammation
D) Recognize pathogen-derived peptides on host MHC surface molecules
E) Express cytoplasmic sensors for detecting pathogen-derived nucleic acids
Recognize pathogen-derived peptides on host MHC surface molecules
2
Both MHC class I and MHC class II molecules are highly polymorphic genes in the human population, with tens to hundreds of different alleles co-existing in the population. This means that a comparison of the MHC protein sequences between two individuals would reveal amino acid differences between one individual and the next. However, these amino acid differences are not randomly distributed along the entire protein, but are clustered in certain locations. The diagram in that most correctly indicates the regions of greatest variability between different MHC proteins (shown by the red highlights) is:
A)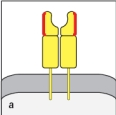
B)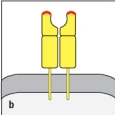
C)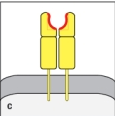
D)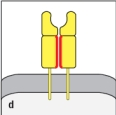
E)
A)

B)

C)

D)

E)


3
MHC class I molecules generally bind peptides that are 8-10 amino acids. Each allelic variant has preferences for the amino acid residues at key anchor positions, but will not bind every possible peptide containing the correct anchor residues.
True
4
The antibody protein has two functional domains, one for antigen binding and a second to confer specific effector functions. These two functional domains are encoded by the antibody light chain and antibody heavy chain polypeptides, respectively.

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
5
MHC genes are the most polymorphic genes in the human genome. This means that, within the population, few individuals share exactly the same sequences for all of their MHC proteins. What deleterious outcome might occur if all humans shared exactly the same sequence for their MHC proteins?

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
6
In Figure Q8), which close-up view of these two V domains has the amino acid sequences most important for antigen-binding highlighted correctly in red?
A)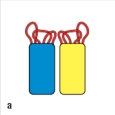
B)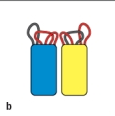
C)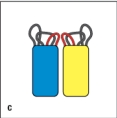
D)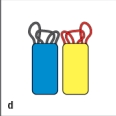
E)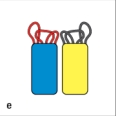

A)

B)

C)

D)

E)


Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
7
When a mixture of different IgG antibody proteins are treated with the enzyme papain, each antibody is cleaved into three roughly equal size fragments. From each original antibody, two of the three fragments are identical to each other, and represent the 'arms' of the antibody 'Y'. These fragments are known as Fab fragments. The third fragment is known as the Fc region, because this fragment will crystallize when purified. The reason a mixture of Fc fragments will crystallize is because:
A) It is the only part of the antibody protein that can easily be purified at the high concentrations needed for crystallization.
B) It has no disulfide bonds holding the domains together, as disulfide bonds will inhibit crystallization.
C) It is the only fragment of the antibody that still has disulfide bonds, so it remains intact during the crystallization process.
D) The Fc fragments of IgG are much more water soluble than the Fab fragments.
E) All Fc fragments generated from a mixture of IgG molecules have the identical amino acid sequence.
A) It is the only part of the antibody protein that can easily be purified at the high concentrations needed for crystallization.
B) It has no disulfide bonds holding the domains together, as disulfide bonds will inhibit crystallization.
C) It is the only fragment of the antibody that still has disulfide bonds, so it remains intact during the crystallization process.
D) The Fc fragments of IgG are much more water soluble than the Fab fragments.
E) All Fc fragments generated from a mixture of IgG molecules have the identical amino acid sequence.

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
8
Early studies analyzing the antibody protein fragments generated after proteolytic cleavage revealed important information about the overall structure of the antibody molecule. Which cleavage pattern (indicated by the red triangles in Figure) yields a fragment that has the same antigen-binding avidity as the intact antibody, but is unable to activate complement after binding to a pathogen?
A)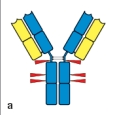
B)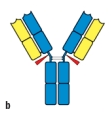
C)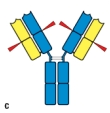
D)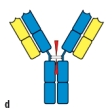
E)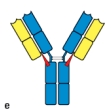
A)

B)

C)

D)

E)


Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
9
Which of the two antibodies shown in Figure are most likely to have the same antigen-binding specificity?
Explain your reasoning.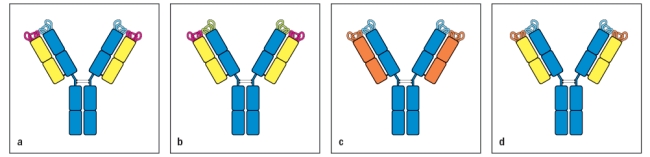
Explain your reasoning.


Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
10
The drawing in Figure shows antibodies bound to repetitive epitopes on the surface of a bacterial pathogen. Even though all of these epitopes are identical, not all of them have antibodies bound to them. 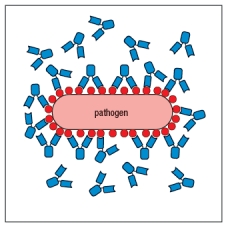 Figure The most likely explanation for this failure of antibodies to bind to every possible epitope on the surface of the pathogen is:
Figure The most likely explanation for this failure of antibodies to bind to every possible epitope on the surface of the pathogen is:
A) There is an insufficient amount of antibody to saturate all the epitopes.
B) The pathogen has an immune evasion strategy to avoid antibody binding to all epitopes.
C) Some of the epitopes cannot bind antibody due to steric hindrance.
D) The antibodies are only able to bind when both antigen-binding sites are engaged on the pathogen surface.
E) The epitopes on the pathogen are not all in the same conformation, so not all will bind the same antibody.
 Figure The most likely explanation for this failure of antibodies to bind to every possible epitope on the surface of the pathogen is:
Figure The most likely explanation for this failure of antibodies to bind to every possible epitope on the surface of the pathogen is:A) There is an insufficient amount of antibody to saturate all the epitopes.
B) The pathogen has an immune evasion strategy to avoid antibody binding to all epitopes.
C) Some of the epitopes cannot bind antibody due to steric hindrance.
D) The antibodies are only able to bind when both antigen-binding sites are engaged on the pathogen surface.
E) The epitopes on the pathogen are not all in the same conformation, so not all will bind the same antibody.

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
11
Individuals or mice with defects in the biochemical pathways needed for loading peptides onto MHC molecules show greatly increased susceptibility to virus infections. Experiments examining the MHC molecules present on the surface of host cells in these individuals would show:
A) Normal numbers of MHC molecules expressed on host cells, but no peptides bound to them.
B) Very low levels of total MHC proteins expressed on the cell surface.
C) Normal numbers of MHC proteins on the surface but all of them bound to self-peptides not pathogen peptides.
D) Very high levels of total MHC proteins expressed on the cell surface.
E) Only virus-infected cells expressing high levels of MHC proteins on the cell surface.
A) Normal numbers of MHC molecules expressed on host cells, but no peptides bound to them.
B) Very low levels of total MHC proteins expressed on the cell surface.
C) Normal numbers of MHC proteins on the surface but all of them bound to self-peptides not pathogen peptides.
D) Very high levels of total MHC proteins expressed on the cell surface.
E) Only virus-infected cells expressing high levels of MHC proteins on the cell surface.

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
12
Amino acid sequence analysis of all of the peptides found in a single IgG antibody would reveal unique peptide sequences totaling ~600-700 amino acids. Using this estimate, the predicted molecular weight of an antibody protein would be ~70-75 kDa. Yet, an intact antibody protein has a molecular weight of ~150 kDa. The explanation for this discrepancy is:
A) IgG antibodies have many more heavy amino acids in them than most other proteins.
B) Each IgG antibody is a complex of two identical light chains and two identical heavy chains.
C) IgG antibodies tend to aggregate together during purification, thereby distorting molecular weight estimates.
D) Each IgG antibody is a complex of four identical polypeptides.
E) IgG antibodies are produced as dimers of two identical IgG monomers.
A) IgG antibodies have many more heavy amino acids in them than most other proteins.
B) Each IgG antibody is a complex of two identical light chains and two identical heavy chains.
C) IgG antibodies tend to aggregate together during purification, thereby distorting molecular weight estimates.
D) Each IgG antibody is a complex of four identical polypeptides.
E) IgG antibodies are produced as dimers of two identical IgG monomers.

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
13
Antibody binding to a pathogen surface is greatly enhanced when both antigen-binding sites of the antibody are engaged at once, a feature known as bivalent binding. It is possible for antibodies to bind bivalently to a wide variety of components on many different pathogen surfaces due to the flexibility in the protein at the hinge region and at the V-C junction.

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
14
Like innate sensors of infections (TLRs, NLRs, RLRs), antibodies frequently recognize nucleic acids of pathogenic organisms.

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
15
Once expressed on the surface of host cells, an MHC protein remains stably associated with its bound peptide for several days. This highly stable peptide binding behavior is important because:
A) It prevents peptide exchanges on the cell surface, ensuring that peptide:MHC complexes are reliable indicators of the proteins present inside that host cell.
B) If the MHC protein lost its peptide it would become unstable, and would be rapidly internalized and degraded.
C) Pathogens would otherwise evade the immune response by making decoy peptides that mimic host cell peptides.
D) Pathogens would be able to evade the T cell response by making proteases that cleave MHC proteins inducing peptide release.
E) Immune responses to infection often induce noxious chemicals that damage surface MHC proteins, and might result in peptide loss.
A) It prevents peptide exchanges on the cell surface, ensuring that peptide:MHC complexes are reliable indicators of the proteins present inside that host cell.
B) If the MHC protein lost its peptide it would become unstable, and would be rapidly internalized and degraded.
C) Pathogens would otherwise evade the immune response by making decoy peptides that mimic host cell peptides.
D) Pathogens would be able to evade the T cell response by making proteases that cleave MHC proteins inducing peptide release.
E) Immune responses to infection often induce noxious chemicals that damage surface MHC proteins, and might result in peptide loss.

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
16
Each immunoglobulin (Ig) domain is composed of a structure known as a ' -sandwich,' which consists of two sheets covalently linked by a disulfide bond. Only a subset of the ~110 amino acids in each domain are required to establish this overall structure, and it is these amino acids that are highly conserved when comparing Ig domains to each other. What might be the advantage of this structure for use as antibody variable domains?

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
17
Some species, like camels, alpacas, and llamas, have evolved variant forms of immunoglobulin proteins that retain the ability to bind to antigens. While overall the antibodies made by these animals are simpler than human or mouse antibodies, an important feature conserved among all of these antibodies is:
A) The presence of both heavy and light chain polypeptides
B) Antigen-binding sites comprised of VH and VL sequences
C) The presence of exactly three constant region domains
D) The presence of two antigen-binding sites per antibody
E) The presence of multiple disulfide bonds linking antibody light chains to heavy chains
A) The presence of both heavy and light chain polypeptides
B) Antigen-binding sites comprised of VH and VL sequences
C) The presence of exactly three constant region domains
D) The presence of two antigen-binding sites per antibody
E) The presence of multiple disulfide bonds linking antibody light chains to heavy chains

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
18
: TCRs are membrane-bound proteins comprised of two polypeptides linked by a disulfide bond. Both polypeptide components of the : TCR are members of the immunoglobulin superfamily, and each of their domains share structural similarity with regions of antibody proteins. However, due to the different functions of TCRs versus antibodies, the overall domain organization of the TCR is not the same as for an antibody. In the cartoon in Figure Q14), describe three features that are incorrect illustrations of the : TCR. 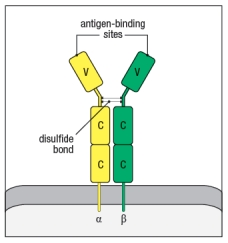


Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
19
The antibody surface involved in antigen binding varies depending on the size and nature of the antigen. This surface can be concave or flat, and sometimes, can have extended protrusions. This is accomplished by:
A) Flexibility in the hinge regions of the antibody allowing rotation of the antigen-binding sites
B) Some antibodies using V region framework sequences instead of the CDRs to bind antigen
C) The ability of different CDR sequences to form many structurally distinct shapes and surfaces
D) The ability of the same heavy chain to pair with different light chains
E) The differential usage of κ versus λ light chains, as κ chains form concave binding sites whereas λ chains make flatter surfaces
A) Flexibility in the hinge regions of the antibody allowing rotation of the antigen-binding sites
B) Some antibodies using V region framework sequences instead of the CDRs to bind antigen
C) The ability of different CDR sequences to form many structurally distinct shapes and surfaces
D) The ability of the same heavy chain to pair with different light chains
E) The differential usage of κ versus λ light chains, as κ chains form concave binding sites whereas λ chains make flatter surfaces

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
20
Antibody heavy and light chain polypeptides consist of repeated domains, each of which is ~110 amino acids and folds up into a compact three-dimensional structure known as an 'immunoglobulin domain.' These immunoglobulin domains are:
A) Mixed and matched between different antibody heavy and light chains to produce variability
B) Always identical to each other within a single antibody heavy chain or light chain polypeptide
C) Always differ in amino acid sequence between different light chain polypeptides in both of the two light chain immunoglobulin domains
D) Similar but not identical in amino acid sequence when comparing the domains in a single heavy chain polypeptide
E) Identical in amino acid sequence for every domain when comparing different antibody heavy chain polypeptides to each other
A) Mixed and matched between different antibody heavy and light chains to produce variability
B) Always identical to each other within a single antibody heavy chain or light chain polypeptide
C) Always differ in amino acid sequence between different light chain polypeptides in both of the two light chain immunoglobulin domains
D) Similar but not identical in amino acid sequence when comparing the domains in a single heavy chain polypeptide
E) Identical in amino acid sequence for every domain when comparing different antibody heavy chain polypeptides to each other

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
21
One striking feature of TCR interactions with peptide:MHC complexes is that amino acid residues in the MHC protein are as important to the TCR binding strength as are amino acid residues in the pathogen-derived peptide. This feature is in contrast to antigen recognition by antibodies, which is a direct interaction that is independent of other host proteins. Based on the different functions of T cells versus antibodies in the adaptive immune response, the fact that TCRs recognize components of both the MHC and the bound peptide exists to:
A) Prevent TCRs from binding only to surface exposed epitopes of native pathogens
B) Prevent immune evasion by a pathogen that has mutated the sequences required for antibody recognition
C) Put constraints on T cell recognition, due to the potentially damaging effector molecules made by activated T cells
D) Ensure that TCRs are focused on recognizing antigens associated with host cells, and not those that are free in solution
E) Ensure that the pathogen has already been destroyed by the host cell before the T cell will recognize it
A) Prevent TCRs from binding only to surface exposed epitopes of native pathogens
B) Prevent immune evasion by a pathogen that has mutated the sequences required for antibody recognition
C) Put constraints on T cell recognition, due to the potentially damaging effector molecules made by activated T cells
D) Ensure that TCRs are focused on recognizing antigens associated with host cells, and not those that are free in solution
E) Ensure that the pathogen has already been destroyed by the host cell before the T cell will recognize it

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
22
: TCRs generally have a binding preference for either peptide:MHC class I or peptide:MHC class II complexes. However, on occasion, one : TCR might actually be able to recognize either class of peptide:MHC complexes. When such an : TCR is expressed on a CD4 T cell, it will only activate its T cell after binding to peptide:MHC class II complexes. Why is this the case?

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
23
The cellular distribution of MHC class I versus MHC class II molecules is quite different, with MHC class II molecules generally expressed on a very limited set of cell types. This is because:
A) It would be detrimental to have CD8 T cells killing macrophages or B cells.
B) CD4 T cells generally secrete cytokines that act on macrophages and B cells.
C) Viruses generally do not infect and replicate in macrophages and B cells.
D) Dendritic cells are more important in stimulating CD4 than CD8 T cell responses.
E) CD4 T cells can only kill macrophages, dendritic cells, and B cells.
A) It would be detrimental to have CD8 T cells killing macrophages or B cells.
B) CD4 T cells generally secrete cytokines that act on macrophages and B cells.
C) Viruses generally do not infect and replicate in macrophages and B cells.
D) Dendritic cells are more important in stimulating CD4 than CD8 T cell responses.
E) CD4 T cells can only kill macrophages, dendritic cells, and B cells.

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
24
Synthesis question: Several vaccines against viral infections are made by isolating purified surface proteins of the viral particle, mixing them with an adjuvant to stimulate an innate immune response, and injecting the mixture into people. Two examples of this are the vaccine against Hepatitis B virus, and the vaccine against Human Papilloma Virus (the 'cervical cancer' vaccine). One interesting property of vaccines of this type (known as 'subunit vaccines') is that there is a requirement for a CD4 T cell response to the vaccine antigen in order to generate antibodies to the innocuous protein in the vaccine. In the case of the Hepatitis B vaccine, the viral protein included in the vaccine is the Hepatitis B surface antigen (HepB-SAg), a protein that is approximately 200 amino acids in length. The graph in Figure shows the data from immunizing individuals with this vaccine, and monitoring their production of protective antibody responses to the viral protein. 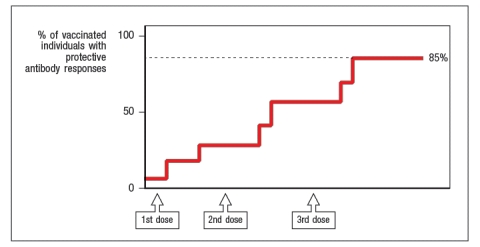
a) What results would be predicted if experiments were performed to examine the CD4 T cell responses to the HepB-SAg in these same individuals? In particular, indicate whether all individuals would show similar responses. Explain your reasoning.
b) Most vaccines, particularly those made by immunizing individuals with purified pathogen components, are designed to elicit robust antibody responses to the immunizing antigens. For vaccines against viral infections (like the HepB vaccine), the immune responses generated are only protective when administered prophylactically, i.e., before the individual is ever exposed to the pathogen. Why is it essential to vaccinate against viral infections before the individual is ever exposed to the virus?

a) What results would be predicted if experiments were performed to examine the CD4 T cell responses to the HepB-SAg in these same individuals? In particular, indicate whether all individuals would show similar responses. Explain your reasoning.
b) Most vaccines, particularly those made by immunizing individuals with purified pathogen components, are designed to elicit robust antibody responses to the immunizing antigens. For vaccines against viral infections (like the HepB vaccine), the immune responses generated are only protective when administered prophylactically, i.e., before the individual is ever exposed to the pathogen. Why is it essential to vaccinate against viral infections before the individual is ever exposed to the virus?

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
25
Synthesis question: For the last five years, the seasonal flu vaccine has contained a mixture of two Influenza A strains and one Influenza B strain. The Influenza A strains were categorized as H1N1 and H3N2 subtypes. This nomenclature refers to the sequences of the two surface glycoproteins on the Influenza A virus particle, the hemaglutinin (H) and the neuraminidase (N). Antibodies specific for these glycoproteins are known to be effective at preventing flu infection. 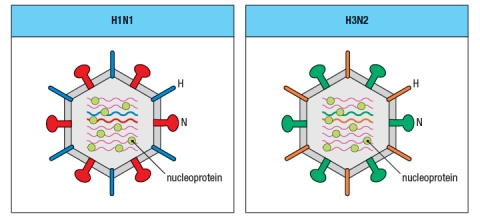
a) The highly pathogenic Asian avian flu causes a fatal infection in about 60% of the individuals infected. The seasonal flu vaccine does not provide protection against this strain of Influenza. Is the highly pathogenic Asian avian flu likely to be an H1N1 or H3N2 strain of influenza? Why or why not?
In the seasonal vaccine, the two strains of Influenza A, H1N1 and H3N2, are both required to provide protection to individuals exposed to one or the other of the viral strains. Currently, research efforts are focused on trying to generate protective CD8 T cell responses to Influenza A, with the goal of generating a 'universal' or broadly neutralizing vaccine that would provide protection against multiple strains of the virus, even those not included in the vaccine.
b) Based on the information provided in the cartoon of the two viral strains shown in Figure , what is the reasoning for expecting CD8 T cell responses to be protective against multiple different strains of Influenza A?
From year to year, the Influenza A strains circulating in the population undergo a process known as 'antigenic drift' in which mutations accumulate in the viral genes, leading to modest changes in the amino acid sequences of the viral proteins. Due to this antigenic drift, different isolates of the H1N1 or H3N2 strains are included in the annual flu vaccine. Shown in Figure are some of the regions of viral proteins that often undergo antigenic drift from year to year.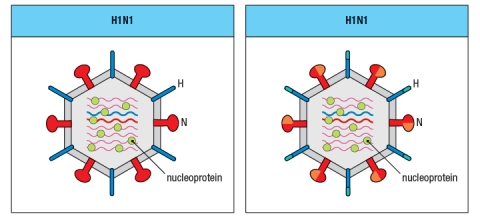
c) What is the minimum number of amino acids that needs to change for a neutralizing antibody to the neuraminidase of the H1N1 strain on the left to no longer bind to the neuraminidase of the H1N1 strain on the right?

a) The highly pathogenic Asian avian flu causes a fatal infection in about 60% of the individuals infected. The seasonal flu vaccine does not provide protection against this strain of Influenza. Is the highly pathogenic Asian avian flu likely to be an H1N1 or H3N2 strain of influenza? Why or why not?
In the seasonal vaccine, the two strains of Influenza A, H1N1 and H3N2, are both required to provide protection to individuals exposed to one or the other of the viral strains. Currently, research efforts are focused on trying to generate protective CD8 T cell responses to Influenza A, with the goal of generating a 'universal' or broadly neutralizing vaccine that would provide protection against multiple strains of the virus, even those not included in the vaccine.
b) Based on the information provided in the cartoon of the two viral strains shown in Figure , what is the reasoning for expecting CD8 T cell responses to be protective against multiple different strains of Influenza A?
From year to year, the Influenza A strains circulating in the population undergo a process known as 'antigenic drift' in which mutations accumulate in the viral genes, leading to modest changes in the amino acid sequences of the viral proteins. Due to this antigenic drift, different isolates of the H1N1 or H3N2 strains are included in the annual flu vaccine. Shown in Figure are some of the regions of viral proteins that often undergo antigenic drift from year to year.

c) What is the minimum number of amino acids that needs to change for a neutralizing antibody to the neuraminidase of the H1N1 strain on the left to no longer bind to the neuraminidase of the H1N1 strain on the right?

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
26
One strategy for vaccine development currently under investigation is the use of pathogen-derived T cell epitopes as a component of the vaccine. For viral pathogens, implementing this strategy involves scanning the predicted amino acid sequences of the viral proteins for likely peptide epitopes that would bind to MHC class I and MHC class II molecules. In addition to the complication of MHC sequence polymorphism in the human population, another complication of this strategy for peptide epitopes that would bind to MHC class II proteins is:
A) The importance of viral proteins containing peptides that are cleaved into 8-10 amino acid long fragments.
B) The ability of viruses to mutate their proteins to avoid MHC anchor residue sequences.
C) The fact that long peptides (>13 amino acids) are rapidly degraded in cells.
D) The fact that MHC class II proteins are intrinsically stable, even in the absence of binding to a peptide.
E) The absence of defined sequence motifs that predict peptide binding to MHC class II molecules.
A) The importance of viral proteins containing peptides that are cleaved into 8-10 amino acid long fragments.
B) The ability of viruses to mutate their proteins to avoid MHC anchor residue sequences.
C) The fact that long peptides (>13 amino acids) are rapidly degraded in cells.
D) The fact that MHC class II proteins are intrinsically stable, even in the absence of binding to a peptide.
E) The absence of defined sequence motifs that predict peptide binding to MHC class II molecules.

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
27
Hepatitis C is a virus that infects hepatocytes, which are non-immune cells of the liver. Currently, patients with chronic Hepatitis C infections are treated with repeated administration of type I interferon, predominantly interferon . One aspect of this treatment that might aid the patient's immune system in clearing this virus infection is:
A) Up-regulation of MHC class I expression levels on hepatocytes
B) Activation of macrophages to produce noxious compounds that might kill the virus
C) Induction of an inflammatory response to promote neutrophil trafficking to the liver
D) Production of TNF- in response to type I interferon leading to vasodilation in the liver
E) Induction of IL-1 and IL-6 leading to the acute phase response
A) Up-regulation of MHC class I expression levels on hepatocytes
B) Activation of macrophages to produce noxious compounds that might kill the virus
C) Induction of an inflammatory response to promote neutrophil trafficking to the liver
D) Production of TNF- in response to type I interferon leading to vasodilation in the liver
E) Induction of IL-1 and IL-6 leading to the acute phase response

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck
28
T cells expressing : TCRs are distinct from those expressing : TCRs in that they do not generally recognize host cell responses to infections or tissue damage; rather they recognize components of the pathogen directly.

Unlock Deck
Unlock for access to all 28 flashcards in this deck.
Unlock Deck
k this deck



