
Molecular Biology of the Gene 7th Edition by Richard Losick, James Watson, Michael Levine, Tamara Baker, Alexander Gann
Edition 7ISBN: 9780321762436
Molecular Biology of the Gene 7th Edition by Richard Losick, James Watson, Michael Levine, Tamara Baker, Alexander Gann
Edition 7ISBN: 9780321762436 Exercise 1
You perform a follow-up experiment to that discussed in the Questions for Chapter 9. Here is a review of the setup for the experiment. You have just discovered two new eukaryotic DNA polymerases and want to learn more about their properties. To begin, you obtain purified protein of each DNA polymerase and perform polymerase processivity assays. You use a short primer that is 32 P-labeled at the 5? end and binds to a circular single-stranded (ssDNA) DNA template (shown below). You complete the following steps to obtain the processivity assay results for DNA polymerase #1 (Pol #1) and DNA polymerase #2 (Pol #2).
1. In the appropriate buffer conditions, you preincubate the primed, circular ssDNA with Pol #1 or Pol #2 for 5 min at 37°C.
2. You add dNTPs and a large excess of ssDNA to initiate the reaction.
3. You allow the reaction to proceed for 10 min, quench the reactions with the addition of SDS, and run the samples on a polyacrylamide gel (which resolves single nucleotides) under denaturing conditions.
You perform a follow-up experiment, and the results are shown below. In this experiment, you separately incubate each DNA polymerase with the accessory proteins (PCNA, RFC, and RPA) and a DNA template that now includes an intrastrand crosslink of two guanines (induced by cisplatin). The cross-link is located immediately adjacent to the 3? end of the primer as indicated by a star (pictured below to the right).
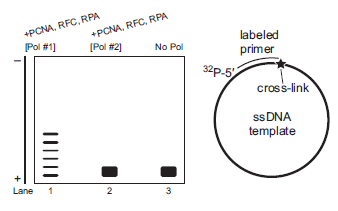
A. Describe the processivity for each DNA polymerase in the presence of a template with a cisplatin-induced intrastrand cross-link.
B. Propose a cellular role for DNA polymerase #1 given this data. Be specific.
1. In the appropriate buffer conditions, you preincubate the primed, circular ssDNA with Pol #1 or Pol #2 for 5 min at 37°C.
2. You add dNTPs and a large excess of ssDNA to initiate the reaction.
3. You allow the reaction to proceed for 10 min, quench the reactions with the addition of SDS, and run the samples on a polyacrylamide gel (which resolves single nucleotides) under denaturing conditions.

You perform a follow-up experiment, and the results are shown below. In this experiment, you separately incubate each DNA polymerase with the accessory proteins (PCNA, RFC, and RPA) and a DNA template that now includes an intrastrand crosslink of two guanines (induced by cisplatin). The cross-link is located immediately adjacent to the 3? end of the primer as indicated by a star (pictured below to the right).

A. Describe the processivity for each DNA polymerase in the presence of a template with a cisplatin-induced intrastrand cross-link.
B. Propose a cellular role for DNA polymerase #1 given this data. Be specific.
Explanation
DNA polymerase #1 (pol #1) is process an...
Molecular Biology of the Gene 7th Edition by Richard Losick, James Watson, Michael Levine, Tamara Baker, Alexander Gann
Why don’t you like this exercise?
Other Minimum 8 character and maximum 255 character
Character 255



