Deck 18: Recombinant Dna and Biotechnology
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question
Question

Unlock Deck
Sign up to unlock the cards in this deck!
Unlock Deck
Unlock Deck
1/243
Play
Full screen (f)
Deck 18: Recombinant Dna and Biotechnology
1
Which enzyme could be used to join two DNA fragments?
A) Reverse transcriptase
B) DNA polymerase
C) DNA recombinase
D) DNA endonuclease
E) DNA ligase
A) Reverse transcriptase
B) DNA polymerase
C) DNA recombinase
D) DNA endonuclease
E) DNA ligase
E
2
The sequence 5ʹ-TCACGT-3ʹ is not a restriction enzyme palindromic recognition sequence.What mutational change would most likely make it into one?
A) Changing T to C at the 5ʹ end
B) Changing T to A at the 5ʹ end
C) Changing T to A at the 3ʹ end
D) Changing T to G at the 3ʹ end
E) No single point mutation can change this sequence into a recognition sequence.
A) Changing T to C at the 5ʹ end
B) Changing T to A at the 5ʹ end
C) Changing T to A at the 3ʹ end
D) Changing T to G at the 3ʹ end
E) No single point mutation can change this sequence into a recognition sequence.
E
3
Which molecule is recombinant DNA?
A) A gene that encodes an enzyme involved in glycolysis that was obtained in the wild from a hybrid between a wolf and a coyote
B) A stretch of DNA that encodes an insulin molecule, created synthetically by chemists in the lab
C) A stretch of DNA that does not code for anything, created by pasting together DNA from a mouse and DNA from a bacterium
D) A stretch of DNA that does not code for anything, from a tomato grown in a garden
E) A gene that codes for a photosynthetic enzyme found in a tomato grown in a garden
A) A gene that encodes an enzyme involved in glycolysis that was obtained in the wild from a hybrid between a wolf and a coyote
B) A stretch of DNA that encodes an insulin molecule, created synthetically by chemists in the lab
C) A stretch of DNA that does not code for anything, created by pasting together DNA from a mouse and DNA from a bacterium
D) A stretch of DNA that does not code for anything, from a tomato grown in a garden
E) A gene that codes for a photosynthetic enzyme found in a tomato grown in a garden
C
4
A restriction enzyme recognizes the sequence 5ʹ-GTCATGAC-3ʹ and makes staggered cuts.Which statement is most likely to be true?
A) Sticky ends should arise because the enzyme makes staggered cuts.
B) Sticky ends should arise because the restriction site starts with a G.
C) Sticky ends should arise because the restriction site is more than six nucleotides long.
D) Blunt ends should arise because the enzyme makes staggered cuts.
E) Blunt ends should arise because the restriction site is more than six nucleotides long.
A) Sticky ends should arise because the enzyme makes staggered cuts.
B) Sticky ends should arise because the restriction site starts with a G.
C) Sticky ends should arise because the restriction site is more than six nucleotides long.
D) Blunt ends should arise because the enzyme makes staggered cuts.
E) Blunt ends should arise because the restriction site is more than six nucleotides long.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
5
Which sequence is a palindromic recognition sequence?
A) 5ʹ-CAATTG-3ʹ
B) 5ʹ-CAATAG-3ʹ
C) 5ʹ-CATTTG-3ʹ
D) 5ʹ-GATTTC-3ʹ
E) 5ʹ-CATCAT-3ʹ
A) 5ʹ-CAATTG-3ʹ
B) 5ʹ-CAATAG-3ʹ
C) 5ʹ-CATTTG-3ʹ
D) 5ʹ-GATTTC-3ʹ
E) 5ʹ-CATCAT-3ʹ

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
6
A molecule results from the joining of DNA from a cow and DNA from a bacterium.It is called
A) a PCR product.
B) complementary DNA.
C) recombinant DNA.
D) a DNA fingerprint.
E) a DNA microarray.
A) a PCR product.
B) complementary DNA.
C) recombinant DNA.
D) a DNA fingerprint.
E) a DNA microarray.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
7
Which enzymes used in biotechnology perform opposite functions?
A) DNA polymerase and DNA ligase
B) RNA polymerase and DNA ligase
C) Restriction endonuclease and DNA ligase
D) Restriction endonuclease and DNA polymerase
E) Reverse transcriptase and restriction endonuclease
A) DNA polymerase and DNA ligase
B) RNA polymerase and DNA ligase
C) Restriction endonuclease and DNA ligase
D) Restriction endonuclease and DNA polymerase
E) Reverse transcriptase and restriction endonuclease

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
8
Refer to the figure showing the recognition sequence for EcoRI.DNA ligase closes the phosphodiester gap between nucleotides that have been cut with restriction enzymes and then reannealed between DNA strands cut with the same restriction enzyme. 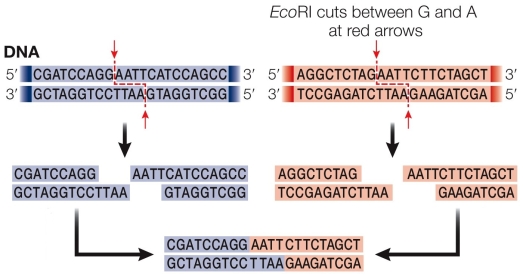 In the figure, the phosphate groups are located on
In the figure, the phosphate groups are located on
A) the guanosine nucleotide located on the top strand and the adenine nucleotide found on the bottom strand.
B) the guanosine nucleotides found on both strands.
C) the adenine nucleotide located on the top strand and the guanosine nucleotide found on the bottom strand.
D) the adenine nucleotides found on both strands.
E) both the adenines and guanosines found on both strands.
 In the figure, the phosphate groups are located on
In the figure, the phosphate groups are located onA) the guanosine nucleotide located on the top strand and the adenine nucleotide found on the bottom strand.
B) the guanosine nucleotides found on both strands.
C) the adenine nucleotide located on the top strand and the guanosine nucleotide found on the bottom strand.
D) the adenine nucleotides found on both strands.
E) both the adenines and guanosines found on both strands.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
9
EcoRI makes staggered cuts when it cleaves DNA, creating single-stranded tails called "sticky ends." These ends can bind to complementary bases.Assuming that concentration of the cut DNA strands is not rate limiting, in order for the complementary ends to base pair, which condition is necessary?
A) The presence of specific helicases
B) Sufficiently high temperature
C) The presence of methyl groups at the end of each DNA strand
D) Sufficiently low temperature
E) Phosphodiester bonds between the complementary DNA strands
A) The presence of specific helicases
B) Sufficiently high temperature
C) The presence of methyl groups at the end of each DNA strand
D) Sufficiently low temperature
E) Phosphodiester bonds between the complementary DNA strands

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
10
Which DNA sequence is most likely a palindromic restriction site? (Note: Only the 5ʹ-to-3ʹ strand is shown.)
A) ATGCTT
B) AGCAGC
C) TAGTAG
D) TCGCGA
E) TGCATA
A) ATGCTT
B) AGCAGC
C) TAGTAG
D) TCGCGA
E) TGCATA

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
11
Which DNA sequences would most likely be recognized by a restriction enzyme that cuts at a palindrome? (Note: Complementary strands are shown.)
A) 5ʹ-AGTCGA-3ʹ and 3ʹ-TCAGCT-5ʹ
B) 5ʹ-GTCTGA-3ʹ and 3ʹ-CAGACT-5ʹ
C) 5ʹ-TAAGGT-3ʹ and 3ʹ-ATTCCA-5ʹ
D) 5ʹ-TAGCTA-3ʹ and 3ʹ-ATCGAT-5ʹ
E) 5ʹ-TCGTGGA-3ʹ and 3ʹ-AGCACCT-5ʹ
A) 5ʹ-AGTCGA-3ʹ and 3ʹ-TCAGCT-5ʹ
B) 5ʹ-GTCTGA-3ʹ and 3ʹ-CAGACT-5ʹ
C) 5ʹ-TAAGGT-3ʹ and 3ʹ-ATTCCA-5ʹ
D) 5ʹ-TAGCTA-3ʹ and 3ʹ-ATCGAT-5ʹ
E) 5ʹ-TCGTGGA-3ʹ and 3ʹ-AGCACCT-5ʹ

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
12
A restriction enzyme recognizes the sequence 5ʹ-CTGCAG-3ʹ and makes cuts between G and C.Which statement is most likely true?
A) Sticky ends should arise because the enzyme cuts in the middle of the sequence.
B) Sticky ends should arise because the restriction site is less than eight nucleotides long.
C) Sticky ends should arise because the restriction site is more than four nucleotides long.
D) Blunt ends should arise because the enzyme cuts once in the middle of the sequence.
E) Blunt ends should arise because the restriction site is more than four nucleotides long
A) Sticky ends should arise because the enzyme cuts in the middle of the sequence.
B) Sticky ends should arise because the restriction site is less than eight nucleotides long.
C) Sticky ends should arise because the restriction site is more than four nucleotides long.
D) Blunt ends should arise because the enzyme cuts once in the middle of the sequence.
E) Blunt ends should arise because the restriction site is more than four nucleotides long

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
13
The sequence 5ʹ-TCATGT-3ʹ is not a restriction enzyme palindromic recognition sequence.What mutational change would most likely make it into one?
A) Changing T to C at the 5ʹ end
B) Changing T to A at the 5ʹ end
C) Changing T to G at the 5ʹ end
D) Changing T to G at the 3ʹ end
E) Changing T to C at the 3ʹ end
A) Changing T to C at the 5ʹ end
B) Changing T to A at the 5ʹ end
C) Changing T to G at the 5ʹ end
D) Changing T to G at the 3ʹ end
E) Changing T to C at the 3ʹ end

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
14
Suppose one restriction enzyme (enzyme A) makes two cuts in a circular DNA molecule, and another restriction enzyme (enzyme B) makes one cut in that same DNA.The cuts are not in the same place, and the cut fragments are not exactly the same size.The restriction digestion is set up on an electrophoresis gel.How many bands should the gel pick up if only enzyme A is used in the restriction digestion?
A) Two
B) Three
C) Four
D) Five
E) Six
A) Two
B) Three
C) Four
D) Five
E) Six

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
15
Suppose a DNA fragment from a human gene has been cut such that it has sticky ends.A DNA fragment from bacteria with sticky ends complementary to those of the human DNA is added to the mix.What would prevent the complementary ends of the human DNA from rejoining, and thus encourage the formation of a recombinant DNA molecule?
A) DNA ligase
B) Restriction endonuclease EcoRI
C) Reverse transcriptase
D) A cloning vector
E) Nothing; the ends could rejoin.
A) DNA ligase
B) Restriction endonuclease EcoRI
C) Reverse transcriptase
D) A cloning vector
E) Nothing; the ends could rejoin.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
16
Which statement about recombinant DNA is true?
A) All restriction endonucleases generate blunt-end fragments.
B) Hydrogen bonds between base pairs are very strong.
C) Blunt ends are exposed bases that can hybridize with complementary sequence fragments.
D) Sticky ends prevent DNA ligase activity.
E) Restriction endonucleases cut both strands of DNA simultaneously.
A) All restriction endonucleases generate blunt-end fragments.
B) Hydrogen bonds between base pairs are very strong.
C) Blunt ends are exposed bases that can hybridize with complementary sequence fragments.
D) Sticky ends prevent DNA ligase activity.
E) Restriction endonucleases cut both strands of DNA simultaneously.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
17
The restriction enzyme AluI produces blunt ends.From this we can conclude that
A) its recognition sequence is less than eight nucleotides long.
B) its recognition sequence is more than four nucleotides long.
C) it cuts once in the recognition sequence.
D) it cuts at two distinct places in the recognition sequence.
E) its recognition sequence is not a palindrome.
A) its recognition sequence is less than eight nucleotides long.
B) its recognition sequence is more than four nucleotides long.
C) it cuts once in the recognition sequence.
D) it cuts at two distinct places in the recognition sequence.
E) its recognition sequence is not a palindrome.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
18
Which activity would be considered cloning of recombinant DNA?
A) Using PCR to produce millions of copies of a specific DNA sequence that came from a human hair for use in forensics
B) Using PCR to produce millions of copies of a specific DNA sequence from a 20,000-year-old wooly mammoth fossil
C) Producing many copies of a gene from a tomato that codes for an enzyme in a yeast cell
D) Using green fluorescent protein as a reporter gene
E) Using a gene that codes for antibiotic resistance as a reporter gene
A) Using PCR to produce millions of copies of a specific DNA sequence that came from a human hair for use in forensics
B) Using PCR to produce millions of copies of a specific DNA sequence from a 20,000-year-old wooly mammoth fossil
C) Producing many copies of a gene from a tomato that codes for an enzyme in a yeast cell
D) Using green fluorescent protein as a reporter gene
E) Using a gene that codes for antibiotic resistance as a reporter gene

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
19
The function of DNA ligase in the generation of recombinant DNA is to
A) cut DNA.
B) replicate DNA.
C) unwind DNA.
D) join DNA fragments.
E) join Okazaki fragments during DNA replication.
A) cut DNA.
B) replicate DNA.
C) unwind DNA.
D) join DNA fragments.
E) join Okazaki fragments during DNA replication.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
20
Which statement regarding recombinant DNA is false?
A) Restriction enzymes can create DNA fragments with "sticky ends."
B) A single restriction enzyme can cut DNA sequences at several different recognition sites.
C) Restriction enzymes cut DNA at palindromic sequences.
D) Restriction enzymes can create DNA fragments with "blunt ends."
E) Complementary "sticky ends" can form hydrogen bonds with each other.
A) Restriction enzymes can create DNA fragments with "sticky ends."
B) A single restriction enzyme can cut DNA sequences at several different recognition sites.
C) Restriction enzymes cut DNA at palindromic sequences.
D) Restriction enzymes can create DNA fragments with "blunt ends."
E) Complementary "sticky ends" can form hydrogen bonds with each other.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
21
Why are plant cells, as opposed to animal cells, particularly useful as hosts for recombinant DNA?
A) Animal cells cannot be cultured.
B) Plant cells lack introns.
C) Plant cells have the ability to become totipotent.
D) Plant cells have reverse transcriptase.
E) Plant cells have plasmids.
A) Animal cells cannot be cultured.
B) Plant cells lack introns.
C) Plant cells have the ability to become totipotent.
D) Plant cells have reverse transcriptase.
E) Plant cells have plasmids.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
22
Which would likely be the worst host for expressing a tomato transgene?
A) Saccharomyces cerevisiae
B) E. coli
C) A mouse cell line
D) A human cell line
E) Another plant species
A) Saccharomyces cerevisiae
B) E. coli
C) A mouse cell line
D) A human cell line
E) Another plant species

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
23
The process of inserting recombinant DNA is considered transfection when
A) the host cells are yeast cells.
B) the host cells are E. coli cells.
C) a selectable marker is used.
D) the vector lacks a replicon.
E) the host cells are mouse tissue culture cells.
A) the host cells are yeast cells.
B) the host cells are E. coli cells.
C) a selectable marker is used.
D) the vector lacks a replicon.
E) the host cells are mouse tissue culture cells.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
24
Bacteria are not particularly useful as hosts in studies of eukaryotic gene expression because they
A) are haploid.
B) have different posttranslational processes.
C) have small genome sizes.
D) contain plasmids.
E) lack genetic markers.
A) are haploid.
B) have different posttranslational processes.
C) have small genome sizes.
D) contain plasmids.
E) lack genetic markers.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
25
To replicate within the cells of a host, recombinant DNA must either _______ the host's genome or contain a(n) _______.Otherwise, the recombinant DNA will not be replicated, because DNA _______ requires specific sequences to bind to DNA.
A) integrate into; origin of replication; polymerase
B) integrate into; vector; polymerase
C) recombine with; origin of replication; ligase
D) recombine with; stop transcription signal; polymerase
E) integrate into; stop transcription signal; ligase
A) integrate into; origin of replication; polymerase
B) integrate into; vector; polymerase
C) recombine with; origin of replication; ligase
D) recombine with; stop transcription signal; polymerase
E) integrate into; stop transcription signal; ligase

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
26
Recombinant DNA technology requires not only getting the DNA into the cell, but also getting it to replicate appropriately.In this process, the specificity of which enzyme can present a challenge?
A) DNA ligase
B) RNA polymerase
C) Replicase
D) Reverse transcriptase
E) DNA polymerase
A) DNA ligase
B) RNA polymerase
C) Replicase
D) Reverse transcriptase
E) DNA polymerase

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
27
In the construction of a transgenic cell, a gene that enables growth on arabinose is added to the vector.This gene is being used as a(n)
A) transformation agent.
B) selectable marker.
C) replicon.
D) cloning vector.
E) expression vector.
A) transformation agent.
B) selectable marker.
C) replicon.
D) cloning vector.
E) expression vector.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
28
In one lab, a gene from tomato that encodes for a protein that increases heat tolerance is transformed into a yeast host.In the same lab, a gene from cows that encodes for a protein that produces a milk protein is transformed into a bacterial host.Which would be considered a transgene?
A) Only the tomato gene in the yeast host
B) Only the cow gene in the bacterial host
C) The tomato gene in its natural tomatoes
D) Both the tomato gene in the yeast host and the cow gene in the tomato host
E) Both the tomato gene in its natural tomatoes and in the yeast host
A) Only the tomato gene in the yeast host
B) Only the cow gene in the bacterial host
C) The tomato gene in its natural tomatoes
D) Both the tomato gene in the yeast host and the cow gene in the tomato host
E) Both the tomato gene in its natural tomatoes and in the yeast host

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
29
Which activity would be considered transfection?
A) Inserting a DNA sequence from cows into a plant host
B) Inserting a DNA sequence from humans into a mouse host
C) Inserting a DNA sequence from humans into a bacterial host
D) Using green fluorescent protein as a reporter gene
E) Using a gene that codes for antibiotic resistance as a reporter gene
A) Inserting a DNA sequence from cows into a plant host
B) Inserting a DNA sequence from humans into a mouse host
C) Inserting a DNA sequence from humans into a bacterial host
D) Using green fluorescent protein as a reporter gene
E) Using a gene that codes for antibiotic resistance as a reporter gene

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
30
Which is not a property of yeast cells that makes them useful as eukaryotic hosts for recombinant DNA studies?
A) Their rapid rate of cell division
B) Their small genome size
C) Their ease of growth in the laboratory
D) The presence of plasmids
E) Their similarity to most other eukaryotic cells
A) Their rapid rate of cell division
B) Their small genome size
C) Their ease of growth in the laboratory
D) The presence of plasmids
E) Their similarity to most other eukaryotic cells

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
31
What changes are typically made to the Ti plasmid when it is used to transform plant cells?
A) Tumor-inducing genes are removed.
B) Tumor-inducing genes are added.
C) Sugar-producing genes are added.
D) Viruses are removed.
E) Viruses are added.
A) Tumor-inducing genes are removed.
B) Tumor-inducing genes are added.
C) Sugar-producing genes are added.
D) Viruses are removed.
E) Viruses are added.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
32
You are choosing between yeast and bacterial cells as hosts for recombinant DNA.Which statement is most true?
A) Possessing plasmids is a feature that would favor bacteria as hosts.
B) Possessing plasmids is a feature that would disfavor bacteria as hosts.
C) Differences between prokaryotes and eukaryotes often tilt the balance toward bacteria as host cells for expressing eukaryotic genes.
D) Bacteria are often favored as hosts because little is known about yeast genetics.
E) Yeasts are often favored as hosts because little is known about bacterial genetics.
A) Possessing plasmids is a feature that would favor bacteria as hosts.
B) Possessing plasmids is a feature that would disfavor bacteria as hosts.
C) Differences between prokaryotes and eukaryotes often tilt the balance toward bacteria as host cells for expressing eukaryotic genes.
D) Bacteria are often favored as hosts because little is known about yeast genetics.
E) Yeasts are often favored as hosts because little is known about bacterial genetics.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
33
A biologist would most likely use the technique of electroporation to
A) separate proteins based on size and charge.
B) knock out a gene.
C) create antisense DNA.
D) measure expression levels of a gene.
E) insert DNA into host cells.
A) separate proteins based on size and charge.
B) knock out a gene.
C) create antisense DNA.
D) measure expression levels of a gene.
E) insert DNA into host cells.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
34
Suppose one restriction enzyme (enzyme A) makes two cuts in a circular DNA molecule, and another restriction enzyme (enzyme B) makes one cut in that same DNA.The cuts are not in the same place, and the cut fragments are not exactly the same size.The restriction digestion is set up on an electrophoresis gel.How many bands should the gel pick up if both enzymes A and B are used in the restriction digestion?
A) Two
B) Three
C) Four
D) Five
E) Six
A) Two
B) Three
C) Four
D) Five
E) Six

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
35
A biologist tries to produce many copies of a particular gene using recombinant DNA technology.This process is called
A) transfection.
B) restriction.
C) cloning.
D) ligating.
E) inducing.
A) transfection.
B) restriction.
C) cloning.
D) ligating.
E) inducing.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
36
You are a researcher interested in getting a pig gene into the replicon of a bacterial host.Which method would you most likely use to get this gene to be part of the replicon?
A) Chemical treatment of host cells to make their outer membranes more permeable
B) Electroporation
C) Use of a gene gun
D) Use of a vector
E) CRISPR-CAS9 technology
A) Chemical treatment of host cells to make their outer membranes more permeable
B) Electroporation
C) Use of a gene gun
D) Use of a vector
E) CRISPR-CAS9 technology

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
37
A plasmid that is to be used in a biotechnology lab is being modified to be more useful as a vector.What is the most likely alteration that will be made?
A) Adding a CAS9 site
B) Adding several unique restriction enzyme recognition sites
C) Electroporation
D) Adding a replicon
E) Adding a genomic library
A) Adding a CAS9 site
B) Adding several unique restriction enzyme recognition sites
C) Electroporation
D) Adding a replicon
E) Adding a genomic library

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
38
In transgenic experiments researchers often make use of electroporation, screen for reporter genes, and insert vectors into the host cell.In which order would they most likely perform these steps?
A) First electroporation, then screen for reporter genes, then insert vectors
B) First electroporation, then insert vectors, then screen for reporter genes
C) First screen for reporter genes, then electroporation, then insert vectors
D) First screen for reporter genes, then insert vectors, then electroporation
E) First insert vectors, then screen for reporter genes, then electroporation
A) First electroporation, then screen for reporter genes, then insert vectors
B) First electroporation, then insert vectors, then screen for reporter genes
C) First screen for reporter genes, then electroporation, then insert vectors
D) First screen for reporter genes, then insert vectors, then electroporation
E) First insert vectors, then screen for reporter genes, then electroporation

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
39
What is the main difference, if any, between transformation and transfection?
A) The nature of the source of the recombinant DNA-it is transformation if the recombinant DNA is only from animals; otherwise it is transfection.
B) The nature of the source of the recombinant DNA-it is transfection if the recombinant DNA is only from animals; otherwise it is transformation.
C) The nature of the host cells-it is transformation if the host cells are animal cells; otherwise it is transfection.
D) The nature of the host cells-it is transfection if the host cells are animal cells; otherwise it is transformation.
E) There is no difference between the two terms.
A) The nature of the source of the recombinant DNA-it is transformation if the recombinant DNA is only from animals; otherwise it is transfection.
B) The nature of the source of the recombinant DNA-it is transfection if the recombinant DNA is only from animals; otherwise it is transformation.
C) The nature of the host cells-it is transformation if the host cells are animal cells; otherwise it is transfection.
D) The nature of the host cells-it is transfection if the host cells are animal cells; otherwise it is transformation.
E) There is no difference between the two terms.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
40
Which characteristic makes plasmids useful as vectors?
A) Many plasmids contain genes that convey antibiotic resistance.
B) Unlike viruses, plasmids do not need to be coaxed to infect cells by artificial means.
C) Plasmids can accommodate a large amount (over 100,000 bp) of DNA.
D) Plasmids use the same origin of replication as eukaryotic cells.
E) All plasmids have only one restriction enzyme recognition site.
A) Many plasmids contain genes that convey antibiotic resistance.
B) Unlike viruses, plasmids do not need to be coaxed to infect cells by artificial means.
C) Plasmids can accommodate a large amount (over 100,000 bp) of DNA.
D) Plasmids use the same origin of replication as eukaryotic cells.
E) All plasmids have only one restriction enzyme recognition site.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
41
Refer to the figure showing an experiment using selectable marker (reporter) genes. 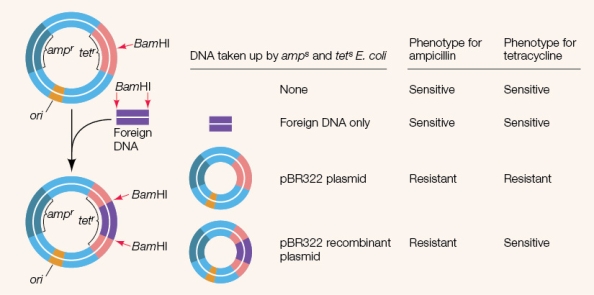 Which statement about the reporter genes used in the experiment is true?
Which statement about the reporter genes used in the experiment is true?
A) Only cells that are resistant to both ampicillin and tetracycline carry the recombinant DNA.
B) The plasmid without incorporated foreign DNA is sensitive to tetracycline and ampicillin.
C) If foreign DNA is successfully incorporated into the plasmid, tetracycline resistance will be disabled.
D) If foreign DNA is successfully incorporated into the plasmid, ampicillin resistance will be disabled.
E) If foreign DNA is successfully incorporated into the plasmid, both tetracycline resistance and ampicillin resistance will be disabled.
 Which statement about the reporter genes used in the experiment is true?
Which statement about the reporter genes used in the experiment is true?A) Only cells that are resistant to both ampicillin and tetracycline carry the recombinant DNA.
B) The plasmid without incorporated foreign DNA is sensitive to tetracycline and ampicillin.
C) If foreign DNA is successfully incorporated into the plasmid, tetracycline resistance will be disabled.
D) If foreign DNA is successfully incorporated into the plasmid, ampicillin resistance will be disabled.
E) If foreign DNA is successfully incorporated into the plasmid, both tetracycline resistance and ampicillin resistance will be disabled.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
42
Suppose a plasmid has genes that confer resistance to both ampicillin and tetracycline.Foreign DNA is inserted at a restriction enzyme recognition site that lies within the ampicillin gene.A cell transformed with the resulting recombinant plasmid should be _______ to ampicillin and _______ to tetracycline.
A) resistant; resistant
B) resistant; sensitive
C) resistant; complementary
D) sensitive; resistant
E) sensitive; complementary
A) resistant; resistant
B) resistant; sensitive
C) resistant; complementary
D) sensitive; resistant
E) sensitive; complementary

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
43
Which statement about selectable marker genes and reporter genes is true?
A) Selectable marker genes are a subset of reporter genes that allow for the survival of bacteria containing the recombinant plasmid.
B) All reporter genes are selectable marker genes.
C) A selectable marker gene cannot be a reporter gene.
D) Reporter genes are antibiotics, whereas selectable marker genes are enzymes.
E) Green fluorescent protein is an example of a product produced from a selectable marker gene but not a reporter gene.
A) Selectable marker genes are a subset of reporter genes that allow for the survival of bacteria containing the recombinant plasmid.
B) All reporter genes are selectable marker genes.
C) A selectable marker gene cannot be a reporter gene.
D) Reporter genes are antibiotics, whereas selectable marker genes are enzymes.
E) Green fluorescent protein is an example of a product produced from a selectable marker gene but not a reporter gene.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
44
cDNA libraries
A) are the same as genomic libraries.
B) include DNA from noncoding sequences.
C) require DNA polymerase for their construction.
D) require reverse transcriptase for their construction.
E) are likely to contain all protein-coding genes.
A) are the same as genomic libraries.
B) include DNA from noncoding sequences.
C) require DNA polymerase for their construction.
D) require reverse transcriptase for their construction.
E) are likely to contain all protein-coding genes.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
45
In which circumstance would you most likely use a virus instead of a plasmid as a vector?
A) When the host cell is bacterial
B) When the host cell is yeast
C) When the transgene is about 5,000 bp long
D) When the transgene is about 15,000 bp long
E) When the transgene is about 50,000 bp long
A) When the host cell is bacterial
B) When the host cell is yeast
C) When the transgene is about 5,000 bp long
D) When the transgene is about 15,000 bp long
E) When the transgene is about 50,000 bp long

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
46
Which statement about genomic and cDNA libraries is true?
A) A genomic library constructed from human brain tissue will be different from a genomic library constructed from human pancreatic tissue.
B) A cDNA library constructed from human brain tissue will be different from a cDNA library constructed from human pancreatic tissue.
C) A cDNA library would be very useful in examining DNA that does not code for mRNA.
D) A genomic library from a tissue will change over time, as opposed to a cDNA library from that same tissue.
E) The construction of a genomic library requires RT-PCR.
A) A genomic library constructed from human brain tissue will be different from a genomic library constructed from human pancreatic tissue.
B) A cDNA library constructed from human brain tissue will be different from a cDNA library constructed from human pancreatic tissue.
C) A cDNA library would be very useful in examining DNA that does not code for mRNA.
D) A genomic library from a tissue will change over time, as opposed to a cDNA library from that same tissue.
E) The construction of a genomic library requires RT-PCR.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
47
Which characteristic would not be useful in a vector?
A) A recognition sequence for a restriction enzyme
B) Large size relative to the host chromosomes
C) The ability to replicate independently inside the host cell
D) A reporter gene
E) An origin of replication
A) A recognition sequence for a restriction enzyme
B) Large size relative to the host chromosomes
C) The ability to replicate independently inside the host cell
D) A reporter gene
E) An origin of replication

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
48
You are interested in determining which genes are being expressed in a muscle cell at a particular time in the development of a chicken.For this, you will most likely use
A) a cDNA library.
B) a genomic library.
C) CRISPR technology.
D) antisense RNA.
E) interference RNA.
A) a cDNA library.
B) a genomic library.
C) CRISPR technology.
D) antisense RNA.
E) interference RNA.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
49
An important vector for the manipulation of plant genes comes from the bacterium Agrobacterium tumefaciens and is called
A) an EcoRI plasmid.
B) (lambda) bacteriophage.
C) BamHI.
D) a Ti plasmid.
E) an ori.
A) an EcoRI plasmid.
B) (lambda) bacteriophage.
C) BamHI.
D) a Ti plasmid.
E) an ori.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
50
A plasmid library and a phage library are constructed from the same genetic material.Which statement is true?
A) The phage library has more clones than the plasmid library because each phage clone contains less DNA.
B) The phage library has fewer clones than the plasmid library because each phage clone contains more DNA.
C) The phage library has fewer clones than the plasmid library because phage libraries do not include nontranscribed DNA.
D) The plasmid library has fewer clones than the phage library because plasmid libraries do not include nontranscribed DNA.
E) The phage library has fewer clones than the plasmid library because phage libraries do not include nontranslated DNA.
A) The phage library has more clones than the plasmid library because each phage clone contains less DNA.
B) The phage library has fewer clones than the plasmid library because each phage clone contains more DNA.
C) The phage library has fewer clones than the plasmid library because phage libraries do not include nontranscribed DNA.
D) The plasmid library has fewer clones than the phage library because plasmid libraries do not include nontranscribed DNA.
E) The phage library has fewer clones than the plasmid library because phage libraries do not include nontranslated DNA.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
51
Which statement concerning a genomic library is true?
A) It comprises the genomic DNA from an organism.
B) It can be made only for eukaryotic organisms.
C) Every colony in the library has only one gene.
D) It can be made only from cDNA.
E) Reverse transcriptase is essential to construct the library.
A) It comprises the genomic DNA from an organism.
B) It can be made only for eukaryotic organisms.
C) Every colony in the library has only one gene.
D) It can be made only from cDNA.
E) Reverse transcriptase is essential to construct the library.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
52
The DNA from the genome of an organism is chopped into numerous fragments that are then inserted into vectors that are taken up by host cells.The genetic information that is present in the resulting colonies is called a genomic
A) collection.
B) library.
C) museum.
D) bank.
E) farm.
A) collection.
B) library.
C) museum.
D) bank.
E) farm.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
53
Biologists engaged in recombinant DNA technology often will cut the vector with two different restriction enzymes that have nearby restriction sites.This is done to
A) increase the size of the vector.
B) increase the probability that the vector will recircularize without the transgene.
C) decrease the probability that the vector will recircularize without the transgene.
D) activate the transgene.
E) inactivate the transgene.
A) increase the size of the vector.
B) increase the probability that the vector will recircularize without the transgene.
C) decrease the probability that the vector will recircularize without the transgene.
D) activate the transgene.
E) inactivate the transgene.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
54
A researcher inserts a DNA segment at the BamHI recognition site within a plasmid; this site is located within the tetracycline resistance gene.This plasmid also has a gene for ampicillin resistance.Following DNA transformation, the researcher must select the bacteria that have taken up the recombinant DNA and not select those that either have taken up the DNA segment only or have taken up nonrecombinant plasmids.In order to do so, the researcher should select the bacteria that
A) are sensitive to both antibiotics.
B) will grow on ampicillin but are sensitive to tetracycline.
C) are resistant to both antibiotics.
D) will grow on tetracycline but are sensitive to ampicillin.
E) grow only on an enriched medium.
A) are sensitive to both antibiotics.
B) will grow on ampicillin but are sensitive to tetracycline.
C) are resistant to both antibiotics.
D) will grow on tetracycline but are sensitive to ampicillin.
E) grow only on an enriched medium.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
55
In which case would use of RT-PCR most likely be the favored method?
A) A researcher is interested in knowing the expression level of one specific gene in brain tissue at a stage of rat development.
B) A researcher is interested in knowing how gene expression patterns across the genome change during acclimation to colder temperatures in a plant species.
C) A researcher is interested in knowing how gene expression patterns across the genome change during development of a fish.
D) A researcher is interested in which genes on the sex chromosomes are silenced during the early stages of meiosis.
E) A researcher is interested in knowing the number of genes in the genome of a puffer fish.
A) A researcher is interested in knowing the expression level of one specific gene in brain tissue at a stage of rat development.
B) A researcher is interested in knowing how gene expression patterns across the genome change during acclimation to colder temperatures in a plant species.
C) A researcher is interested in knowing how gene expression patterns across the genome change during development of a fish.
D) A researcher is interested in which genes on the sex chromosomes are silenced during the early stages of meiosis.
E) A researcher is interested in knowing the number of genes in the genome of a puffer fish.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
56
Which technique makes use of reverse transcriptase?
A) Southern blotting
B) Construction of a gene library
C) Construction of a cDNA library
D) Gene knockout study
E) Pharming
A) Southern blotting
B) Construction of a gene library
C) Construction of a cDNA library
D) Gene knockout study
E) Pharming

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
57
Refer to the figure showing an experiment using selectable marker (reporter) genes. 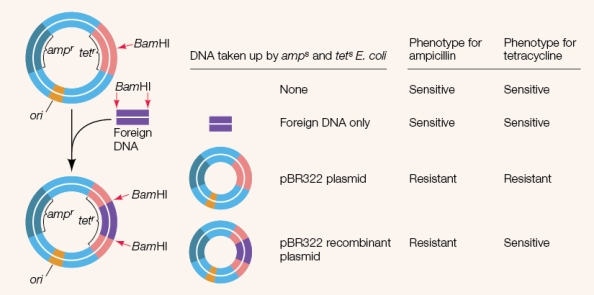 Which conclusion is supported by this experiment?
Which conclusion is supported by this experiment?
A) E coli can be either sensitive or resistant to specific antibiotics.
B) Antibiotic resistance may be conferred to the bacterium by genes in plasmids.
C) Foreign DNA does not transform the bacterium.
D) Transformed bacteria that contain foreign DNA may be identified by insertion of the foreign DNA into an antibiotic resistance gene.
E) Antibiotic resistance genes are good selectable markers.
 Which conclusion is supported by this experiment?
Which conclusion is supported by this experiment?A) E coli can be either sensitive or resistant to specific antibiotics.
B) Antibiotic resistance may be conferred to the bacterium by genes in plasmids.
C) Foreign DNA does not transform the bacterium.
D) Transformed bacteria that contain foreign DNA may be identified by insertion of the foreign DNA into an antibiotic resistance gene.
E) Antibiotic resistance genes are good selectable markers.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
58
The plant disease called crown gall is caused by the
A) insertion into plant cells of a T DNA element carried on the Ti plasmid.
B) transcription of the Ti plasmid in the plant cells.
C) translation of the Ti plasmid in the plant cells.
D) rampant multiplication of Agrobacterium tumefaciens bacteria within the plant.
E) transfer of bacterial genomes into the plant cell genome.
A) insertion into plant cells of a T DNA element carried on the Ti plasmid.
B) transcription of the Ti plasmid in the plant cells.
C) translation of the Ti plasmid in the plant cells.
D) rampant multiplication of Agrobacterium tumefaciens bacteria within the plant.
E) transfer of bacterial genomes into the plant cell genome.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
59
What is the major difference between using green fluorescent protein and using an antibiotic resistance gene as a reporter gene?
A) The green fluorescent protein can be used only when the host cell is a plant cell because it relies on the process of photosynthesis.
B) The antibiotic resistance gene can be used only when the host cell is an animal.
C) The green fluorescent protein can be used only in transfection, not in transformation.
D) With the green fluorescent protein, no cells are killed in the selection process.
E) Homologous recombination will work only with the green fluorescent protein.
A) The green fluorescent protein can be used only when the host cell is a plant cell because it relies on the process of photosynthesis.
B) The antibiotic resistance gene can be used only when the host cell is an animal.
C) The green fluorescent protein can be used only in transfection, not in transformation.
D) With the green fluorescent protein, no cells are killed in the selection process.
E) Homologous recombination will work only with the green fluorescent protein.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
60
What was the original source of the green fluorescent protein?
A) Green algae
B) Arabidopsis thaliana
C) Yeast
D) A firefly
E) A jellyfish
A) Green algae
B) Arabidopsis thaliana
C) Yeast
D) A firefly
E) A jellyfish

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
61
A bacterium that has the CRISPR-CAS9 system has a mutation in one of its spacer sequences.What is the most likely result?
A) The bacterium will be unable to recognize particular viruses.
B) The bacterium will be able to recognize viruses but will be unable to cut them.
C) The bacterium will overexpress all of its genes.
D) The bacterium will overexpress particular genes.
E) The bacterium will underexpress particular genes.
A) The bacterium will be unable to recognize particular viruses.
B) The bacterium will be able to recognize viruses but will be unable to cut them.
C) The bacterium will overexpress all of its genes.
D) The bacterium will overexpress particular genes.
E) The bacterium will underexpress particular genes.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
62
A biologist used microarrays to examine how gene expression in a fish changes with temperature.Refer to the table showing the expression levels of four (of several thousand) genes at varying temperatures.Values for expression have been normalized such that the expression at 15°C for any particular gene is always 100. 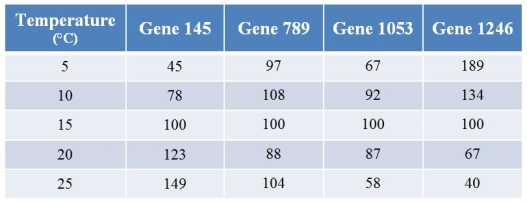 Which two genes show the most opposite patterns of expression as temperature changes?
Which two genes show the most opposite patterns of expression as temperature changes?
A) Genes 145 and 789
B) Genes 145 and 1246
C) Genes 789 and 1053
D) Genes 789 and 1246
E) Genes 1053 and 1246
 Which two genes show the most opposite patterns of expression as temperature changes?
Which two genes show the most opposite patterns of expression as temperature changes?A) Genes 145 and 789
B) Genes 145 and 1246
C) Genes 789 and 1053
D) Genes 789 and 1246
E) Genes 1053 and 1246

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
63
A bacterium that has the CRISPR-CAS9 system has a CAS9 that has much reduced activity.What is the most likely result?
A) The bacterium will be able to recognize viruses but will be unable to cut them.
B) The bacterium will be unable to recognize particular viruses.
C) The bacterium will cut its own DNA inappropriately.
D) The bacterium will overexpress particular genes.
E) The bacterium will underexpress particular genes.
A) The bacterium will be able to recognize viruses but will be unable to cut them.
B) The bacterium will be unable to recognize particular viruses.
C) The bacterium will cut its own DNA inappropriately.
D) The bacterium will overexpress particular genes.
E) The bacterium will underexpress particular genes.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
64
What is the advantage of RNAi over antisense RNA in blocking translation?
A) RNAi can be used with DNA chips.
B) Only RNAi can be used to test cause-and-effect relationships.
C) RNAi is more stable than antisense RNA.
D) RNAi is single-stranded and thus easier to produce.
E) RNAi blocks transcription.
A) RNAi can be used with DNA chips.
B) Only RNAi can be used to test cause-and-effect relationships.
C) RNAi is more stable than antisense RNA.
D) RNAi is single-stranded and thus easier to produce.
E) RNAi blocks transcription.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
65
What role do spacers play in the CRISPR-CAS9 system found in many bacteria?
A) Destroying natural enemies
B) DNA repair
C) Increasing gene expression
D) Decreasing gene expression
E) Identification of natural enemies
A) Destroying natural enemies
B) DNA repair
C) Increasing gene expression
D) Decreasing gene expression
E) Identification of natural enemies

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
66
A biologist used microarrays to examine how gene expression in a fish changes with temperature.Refer to the table showing the expression levels of four (of several thousand) genes at varying temperatures.Values for expression have been normalized such that the expression at 15°C for any particular gene is always 100. 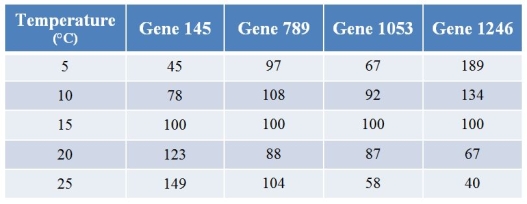 The product of gene 1246 is an inhibitor of a pigment that makes fish red.Thus, all other things being equal and assuming that a 25 percent change in expression level is sufficient for a color change, what should be expected?
The product of gene 1246 is an inhibitor of a pigment that makes fish red.Thus, all other things being equal and assuming that a 25 percent change in expression level is sufficient for a color change, what should be expected?
A) The fish would be the most red at just the highest temperatures.
B) The fish would be the most red at just the lowest temperatures.
C) The fish would be the most red at intermediate temperatures, and less red at the extremes.
D) The fish would be less red at intermediate temperatures, and redder at the extremes.
E) Fish color should not vary much with temperature.
 The product of gene 1246 is an inhibitor of a pigment that makes fish red.Thus, all other things being equal and assuming that a 25 percent change in expression level is sufficient for a color change, what should be expected?
The product of gene 1246 is an inhibitor of a pigment that makes fish red.Thus, all other things being equal and assuming that a 25 percent change in expression level is sufficient for a color change, what should be expected?A) The fish would be the most red at just the highest temperatures.
B) The fish would be the most red at just the lowest temperatures.
C) The fish would be the most red at intermediate temperatures, and less red at the extremes.
D) The fish would be less red at intermediate temperatures, and redder at the extremes.
E) Fish color should not vary much with temperature.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
67
DNA microarrays
A) can be used to compare gene expression levels in different tissues.
B) are used to analyze genomic DNA.
C) use oligonucleotides as probes.
D) probe noncoding regions of DNA.
E) detect proteins that are translated at different times or in different tissues.
A) can be used to compare gene expression levels in different tissues.
B) are used to analyze genomic DNA.
C) use oligonucleotides as probes.
D) probe noncoding regions of DNA.
E) detect proteins that are translated at different times or in different tissues.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
68
Sequences of a mammal find an association between variants at a SNP in an enzyme and coat color.Biologists do not know whether the coat color differences are actually due to that SNP or are just correlated with the SNP and are instead due to other differences.What would be the most appropriate way to find out whether the association between the SNP and coat color was causal?
A) Use CRISPR technology to alter the expression of the gene.
B) Use CRISPR technology to mutate the SNP.
C) Use RNAi to alter expression of the gene.
D) Use RNAi to mutate the SNP.
E) Use antisense RNA to mutate the SNP.
A) Use CRISPR technology to alter the expression of the gene.
B) Use CRISPR technology to mutate the SNP.
C) Use RNAi to alter expression of the gene.
D) Use RNAi to mutate the SNP.
E) Use antisense RNA to mutate the SNP.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
69
Refer to the table showing results from an experiment in which researchers used CRISPR technology to knock out various genes from a plant.They then examined plant stem height (in centimeters, with calculated means) and seed production (number of seeds, with calculated means) with each treatment.(Note: The "x" marks genes that have been knocked out.) 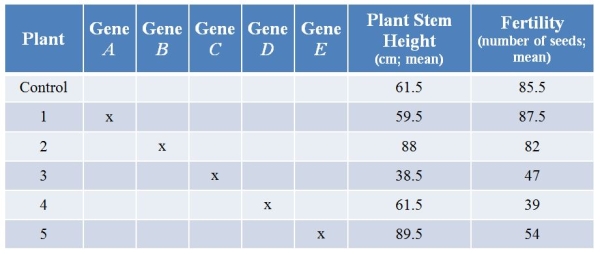 What is the most likely role of the product of gene B?
What is the most likely role of the product of gene B?
A) It is a stimulator of both stem growth and seed production.
B) It is a stimulator of stem growth and an inhibitor of seed production.
C) It is an inhibitor of stem growth, and it stimulates seed production.
D) It is an inhibitor of stem growth and has no discernable effect on seed production.
E) It has no discernable effect on either trait.
 What is the most likely role of the product of gene B?
What is the most likely role of the product of gene B?A) It is a stimulator of both stem growth and seed production.
B) It is a stimulator of stem growth and an inhibitor of seed production.
C) It is an inhibitor of stem growth, and it stimulates seed production.
D) It is an inhibitor of stem growth and has no discernable effect on seed production.
E) It has no discernable effect on either trait.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
70
Biologists examining the plant gene LEAFY's effect on expression patterns of genes overexpressed and underexpressed LEAFY.They then performed microarray analysis to examine the effects of changes of LEAFY expression on the expression of other genes in the genome.Refer to the table showing the results for four different genes with LEAFY underexpressed and overexpressed, and the control.The expression levels are normalized such that they are 100 for each gene in control conditions. 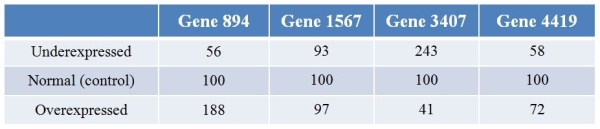 Which pattern is exhibited by gene 3407?
Which pattern is exhibited by gene 3407?
A) As expression of LEAFY increases, the expression of gene 3407 also increases.
B) As expression of LEAFY increases, the expression of gene 3407 decreases readily.
C) Gene 3407 is most expressed when LEAFY is normally expressed; its expression decreases as LEAFY is either overexpressed or underexpressed.
D) Gene 3407 is least expressed when LEAFY is normally expressed; its expression increases as LEAFY is either overexpressed or underexpressed.
E) LEAFY has little or no effect on the expression of gene 3407.
 Which pattern is exhibited by gene 3407?
Which pattern is exhibited by gene 3407?A) As expression of LEAFY increases, the expression of gene 3407 also increases.
B) As expression of LEAFY increases, the expression of gene 3407 decreases readily.
C) Gene 3407 is most expressed when LEAFY is normally expressed; its expression decreases as LEAFY is either overexpressed or underexpressed.
D) Gene 3407 is least expressed when LEAFY is normally expressed; its expression increases as LEAFY is either overexpressed or underexpressed.
E) LEAFY has little or no effect on the expression of gene 3407.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
71
A biologist used microarrays to examine how gene expression in a fish changes with temperature.Refer to the table showing the expression levels of four (of several thousand) genes at varying temperatures.Values for expression have been normalized such that the expression at 15°C for any particular gene is always 100. 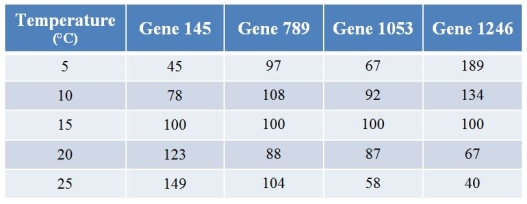 What best describes the pattern shown by gene 1053?
What best describes the pattern shown by gene 1053?
A) Its expression steadily increases with temperature.
B) Its expression steadily decreases with temperature.
C) Its expression first increases and then decreases as temperature increases.
D) Its expression first decreases and then increases as temperature increases.
E) Temperature has little or no effect on its expression.
 What best describes the pattern shown by gene 1053?
What best describes the pattern shown by gene 1053?A) Its expression steadily increases with temperature.
B) Its expression steadily decreases with temperature.
C) Its expression first increases and then decreases as temperature increases.
D) Its expression first decreases and then increases as temperature increases.
E) Temperature has little or no effect on its expression.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
72
Variation at a specific gene is known to affect metabolism in mice, but it is not clear which of the nucleotides is responsible for the changes.Which technique would be most relevant for revealing which of the nucleotides affects metabolism?
A) Microarrays
B) Antisense RNA
C) RNAi
D) Artificial synthesis
E) CRISPR
A) Microarrays
B) Antisense RNA
C) RNAi
D) Artificial synthesis
E) CRISPR

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
73
It is suspected that the gene DEFT regulates the expression of many other genes.How could one test this?
A) Knock out different genes using CRISPR, then see how DEFT is expressed using microarrays.
B) Knock out different genes using CRISPR, then see how DEFT is expressed using RNAi.
C) Use CRISPR to knock DEFT out, then use microarrays to examine how genes are expressed with and without DEFT expression.
D) Use CRISPR to knock DEFT out, then use RNAi to examine how genes are expressed with and without DEFT expression.
E) Use microarrays to knock out DEFT, then use RT-PCR to see how genes are expressed with and without DEFT expression.
A) Knock out different genes using CRISPR, then see how DEFT is expressed using microarrays.
B) Knock out different genes using CRISPR, then see how DEFT is expressed using RNAi.
C) Use CRISPR to knock DEFT out, then use microarrays to examine how genes are expressed with and without DEFT expression.
D) Use CRISPR to knock DEFT out, then use RNAi to examine how genes are expressed with and without DEFT expression.
E) Use microarrays to knock out DEFT, then use RT-PCR to see how genes are expressed with and without DEFT expression.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
74
Refer to the table showing results from an experiment in which researchers used CRISPR technology to knock out various genes from a plant.They then examined plant stem height (in centimeters, with calculated means) and seed production (number of seeds, with calculated means) with each treatment.(Note: The "x" marks genes that have been knocked out.) 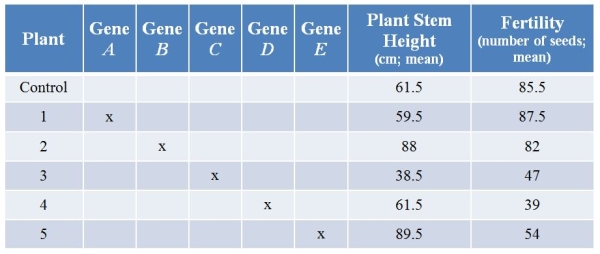 What is the most likely role of the product of gene A?
What is the most likely role of the product of gene A?
A) It is a stimulator of both stem growth and seed production.
B) It is a stimulator of stem growth and an inhibitor of seed production.
C) It is an inhibitor of stem growth, and it stimulates seed production.
D) It is an inhibitor of stem growth and has no discernable effect on seed production.
E) It has no discernable effect on either trait.
 What is the most likely role of the product of gene A?
What is the most likely role of the product of gene A?A) It is a stimulator of both stem growth and seed production.
B) It is a stimulator of stem growth and an inhibitor of seed production.
C) It is an inhibitor of stem growth, and it stimulates seed production.
D) It is an inhibitor of stem growth and has no discernable effect on seed production.
E) It has no discernable effect on either trait.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
75
Which statement about antisense RNA and RNAi is true?
A) Antisense RNA and RNAi both block translation, but antisense RNA is generally preferred.
B) Antisense RNA and RNAi both block translation, but RNAi is generally preferred.
C) Antisense RNA and RNAi both block transcription, but antisense RNA is generally preferred.
D) Antisense RNA and RNAi both block transcription, but RNAi is generally preferred.
E) Antisense RNA and RNAi are different names for the same technique.
A) Antisense RNA and RNAi both block translation, but antisense RNA is generally preferred.
B) Antisense RNA and RNAi both block translation, but RNAi is generally preferred.
C) Antisense RNA and RNAi both block transcription, but antisense RNA is generally preferred.
D) Antisense RNA and RNAi both block transcription, but RNAi is generally preferred.
E) Antisense RNA and RNAi are different names for the same technique.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
76
Which technology is used to examine the patterns of gene expression for a large number of genes simultaneously?
A) Antisense RNA
B) RNA interference
C) DNA microarray
D) PCR
E) CRISPR technology
A) Antisense RNA
B) RNA interference
C) DNA microarray
D) PCR
E) CRISPR technology

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
77
Refer to the table showing results from an experiment in which researchers used CRISPR technology to knock out various genes from a plant.They then examined plant stem height (in centimeters, with calculated means) and seed production (number of seeds, with calculated means) with each treatment.(Note: The "x" marks genes that have been knocked out.) 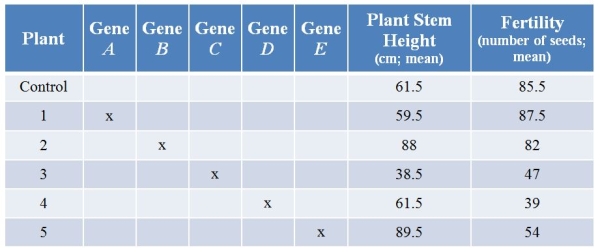 Knocking out which gene had effects only on fertility?
Knocking out which gene had effects only on fertility?
A) Gene A
B) Gene B
C) Gene C
D) Gene D
E) Gene E
 Knocking out which gene had effects only on fertility?
Knocking out which gene had effects only on fertility?A) Gene A
B) Gene B
C) Gene C
D) Gene D
E) Gene E

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
78
In which circumstance would microarrays most likely be used?
A) A biologist is interested in determining the effect of a particular SNP of a gene on flowering time of a plant.
B) A lab is studying a gene and its effects on mouse behavior; they want to know whether knocking out that gene alters food-seeking behavior.
C) A developmental biologist is examining genome-wide patterns of gene expression in different tissues of a fish in response to cold stress.
D) An evolutionary biologist is interested in whether genes that are on the X chromosome evolve faster than those that are autosomal.
E) A lab wants to make large quantities of a pharmaceutical drug using cows.
A) A biologist is interested in determining the effect of a particular SNP of a gene on flowering time of a plant.
B) A lab is studying a gene and its effects on mouse behavior; they want to know whether knocking out that gene alters food-seeking behavior.
C) A developmental biologist is examining genome-wide patterns of gene expression in different tissues of a fish in response to cold stress.
D) An evolutionary biologist is interested in whether genes that are on the X chromosome evolve faster than those that are autosomal.
E) A lab wants to make large quantities of a pharmaceutical drug using cows.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
79
Refer to the table showing results from an experiment in which researchers used CRISPR technology to knock out various genes from a plant.They then examined plant stem height (in centimeters, with calculated means) and seed production (number of seeds, with calculated means) with each treatment.(Note: The "x" marks genes that have been knocked out.) 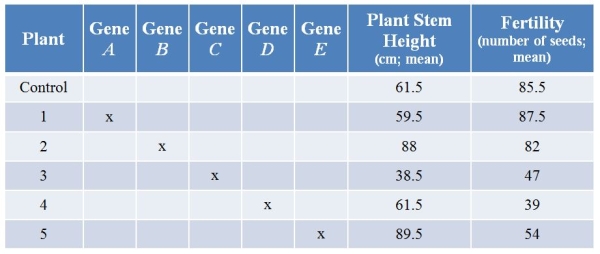 What is the most likely role of the product of gene C?
What is the most likely role of the product of gene C?
A) It is a stimulator of both stem growth and seed production.
B) It is a stimulator of stem growth and an inhibitor of seed production.
C) It is an inhibitor of stem growth, and it stimulates seed production.
D) It is an inhibitor of stem growth and has no discernable effect on seed production.
E) It has no discernable effect on either trait.
 What is the most likely role of the product of gene C?
What is the most likely role of the product of gene C?A) It is a stimulator of both stem growth and seed production.
B) It is a stimulator of stem growth and an inhibitor of seed production.
C) It is an inhibitor of stem growth, and it stimulates seed production.
D) It is an inhibitor of stem growth and has no discernable effect on seed production.
E) It has no discernable effect on either trait.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck
80
A biologist used microarrays to examine how gene expression in a fish changes with temperature.Refer to the table showing the expression levels of four (of several thousand) genes at varying temperatures.Values for expression have been normalized such that the expression at 15°C for any particular gene is always 100. 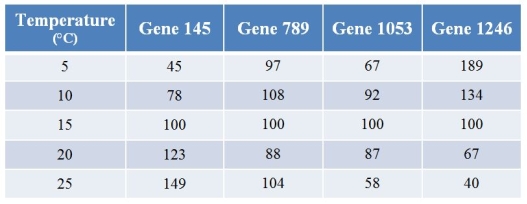 What best describes the pattern shown by gene 789?
What best describes the pattern shown by gene 789?
A) Its expression steadily increases with temperature.
B) Its expression steadily decreases with temperature.
C) Its expression first increases and then decreases as temperature increases.
D) Its expression first decreases and then increases as temperature increases.
E) Temperature has little or no effect on its expression.
 What best describes the pattern shown by gene 789?
What best describes the pattern shown by gene 789?A) Its expression steadily increases with temperature.
B) Its expression steadily decreases with temperature.
C) Its expression first increases and then decreases as temperature increases.
D) Its expression first decreases and then increases as temperature increases.
E) Temperature has little or no effect on its expression.

Unlock Deck
Unlock for access to all 243 flashcards in this deck.
Unlock Deck
k this deck



