You are trying to identify the peroxisome-targeting sequence in the thiolase enzyme in yeast.The thiolase enzyme normally resides in the peroxisome and therefore must contain amino acid sequences that are used to target the enzyme for import into the peroxisome.To identify the targeting sequences, you create a set of hybrid genes that encode fusion proteins containing part of the thiolase protein fused to another protein, histidinol dehydrogenase (HDH).HDH is a cytosolic enzyme required for the synthesis of the amino acid histidine and cannot function if it is localized in the peroxisome.You genetically engineer a series of yeast cells to express these fusion proteins instead of their own versions of these enzymes.If the fusion proteins are imported into the peroxisome, the HDH portion of the protein cannot function and the yeast cells cannot grow on a medium lacking histidine.You obtain the results shown in Figure 15-5. 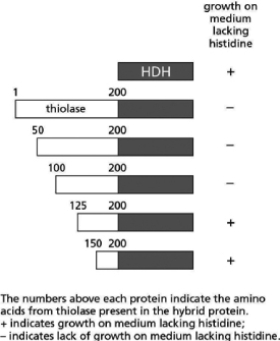
Figure 15-5
What region of the thiolase protein contains the peroxisomal targeting sequence? Explain your answer.
Correct Answer:
Verified
View Answer
Unlock this answer now
Get Access to more Verified Answers free of charge
Q43: For each of the following sentences,
Q44: A gene regulatory protein, A, contains a
Q45: v-SNAREs and t-SNAREs mediate the recognition of
Q46: For each of the following sentences,
Q47: The interior of the organelle contains ribosomes.
How
Q49: Using genetic engineering techniques, you have created
Q50: For each of the following sentences,
Q51: In a cell capable of regulated secretion,
Q52: What would happen in each of the
Q53: Name a type of protein modification that
Unlock this Answer For Free Now!
View this answer and more for free by performing one of the following actions

Scan the QR code to install the App and get 2 free unlocks

Unlock quizzes for free by uploading documents