Passage
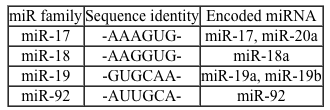
Overexpression of the miR-17~92 gene cluster occurs in many human cancers involving c-Myc, a gene coding for a transcription factor that regulates expression of proteins such as p21, an inhibitor of cell cycle progression. miR-17~92 encodes six distinct microRNAs (miRNAs) that can be divided into four families based on sequence identity at positions 2-7 (Table 1) .Table 1 Sequence Identities of miRNA Families Encoded by the mir-17∼92 Gene Cluster
 Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ) , which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ) . As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92) .
Experiment 1Two independent, diffuse B-cell lymphoma cell lines were used to study the functional relationship between miR-17~92 transcripts and c-Myc in cancer pathogenesis. The first cell line was derived from Eμ-Myc mice (Eμ) , which overexpress a c-Myc transgene under the control of a B-cell-specific enhancer that induces early development of B-cell lymphoma. The second cell line was derived from Eμ-Myc mice carrying a miR-17∼92 knockout allele (Δ/Δ) . As shown in Figure 1, researchers measured the growth of both cell lines, as well as that of Δ/Δ cells infected with a retrovirus expressing the entire miR-17∼92 cluster (Δ/Δ + miR-17∼92) .
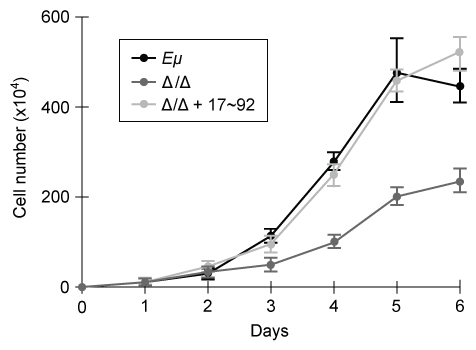 Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation) Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2) .
Figure 1 Growth curves of Eμ cells, Δ/Δ cells, and Δ/Δ + miR-17∼92 cells (error bars = standard deviation) Experiment 2To assess the relative contribution of each miRNA to the oncogenic ability of the miR-17∼92 cluster, Δ/Δ cell lines were transduced with an empty vector or with miR-17∼92 mutant allele constructs that were engineered to lack expression of miRNA from at least one of the four families. In each of the constructs, the family that was knocked out of the vector is indicated by the Δ symbol. After verifying that the transduced cells correctly expressed the desired miRNA, the fraction of apoptotic cells in each line was measured (Figure 2) .
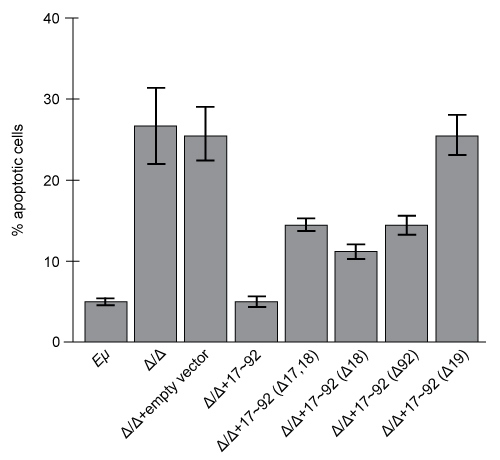 Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Figure 2 Percentage of apoptotic cells in Eμ cells and Δ/Δ cells transduced with the indicated miR-17~92 construct (error bars = standard deviation)
Adapted from Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24) :2806-11.
-If changes in p21 expression contribute to the development of B-cell lymphoma in Eμ-Myc mice, what is the expected result of real-time PCR analysis of c-Myc and p21 expression in wild-type (wt) and Eμ-Myc (Eμ) mice? (Note: mRNA levels are standardized to the expression of ubiquitin (Ub) , which is not regulated by c-Myc.)
A) 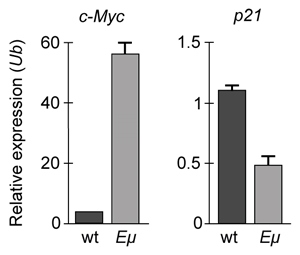
B) 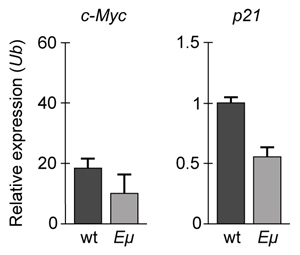
C) 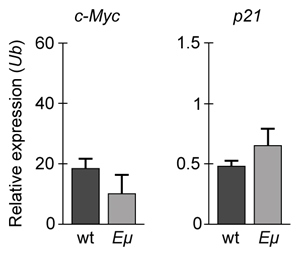
D) 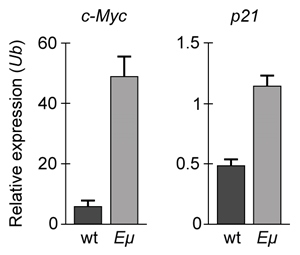
Correct Answer:
Verified
Q48: Passage
Alzheimer disease (AD) is a progressive condition
Q49: Passage
Alzheimer disease (AD) is a progressive condition
Q50: Passage
Alzheimer disease (AD) is a progressive condition
Q51: Passage
Overexpression of the miR-17~92 gene cluster occurs
Q52: Passage
Alzheimer disease (AD) is a progressive condition
Q54: Passage
The lac operon encodes the proteins that
Q55: Passage
Overexpression of the miR-17~92 gene cluster occurs
Q56: Passage
The lac operon encodes the proteins that
Q57: Passage
The lac operon encodes the proteins that
Q58: Passage
Alzheimer disease (AD) is a progressive condition
Unlock this Answer For Free Now!
View this answer and more for free by performing one of the following actions

Scan the QR code to install the App and get 2 free unlocks

Unlock quizzes for free by uploading documents