Morphologically, Species A is very similar to four other species, B-E. Yet the nucleotide sequence deep within an intron in a gene shared by all five of these eukaryotic species is quite different in Species A compared to that of the other four species when one studies the nucleotides present at each position.
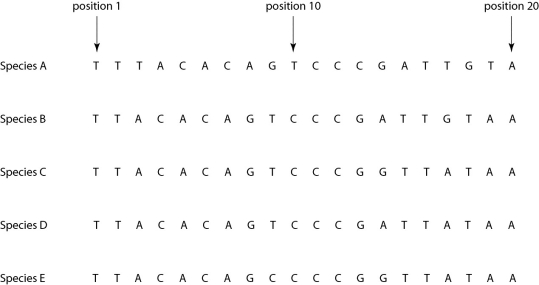 Figure 26.4
Figure 26.4
-If the sequence of Species A differs from that of the other four species due to simple misalignment, then what should the computer software find when it compares the sequence of Species A to those of the other four species?
A) The nucleotide at position 1 should be different in Species A, but the same in species B-E.
B) The nucleotide sequence of Species A should have long sequences that are nearly identical to those of the other species, but offset in terms of position number.
C) The sequences of species B-E, though different from that of Species A, should be identical to each other, without exception.
D) If the software compares, not nucleotide sequence, but rather the amino acid sequence of the actual protein product, then the amino acid sequences of species B-E should be similar to each other, but very different from that of Species A.
E) Computer software is useless in determining sequences of introns; it can only be used with exons.
Correct Answer:
Verified
Q33: The most important feature that permits a
Q45: The following questions refer to the
Q46: The following questions refer to the
Q47: The following questions refer to the
Q48: Neutral theory proposes that
A)molecular clocks are more
Q49: The following questions refer to the
Q51: The following questions refer to the
Q52: The following questions refer to the
Q54: The following questions refer to the
Q55: The following questions refer to the
Unlock this Answer For Free Now!
View this answer and more for free by performing one of the following actions

Scan the QR code to install the App and get 2 free unlocks

Unlock quizzes for free by uploading documents