Passage
The Bloom syndrome helicase (BLM) transcript is catalytically inactive in Bloom syndrome (BS) , a genetic disorder that leads to genomic instability characterized by excessive somatic recombination events. Research studies have shown that BLM may be involved in DNA synthesis repair of stalled replication forks during replication stress (arrest) . To investigate this further, an experiment was conducted using cell lines derived from a patient with BS. One set of cells differed only by BLM status: PSNF5 (BLM+, vector with BLM cDNA transfected) and PSNG13 (BLM−, empty vector transfected) . Additional mutations, BLM-K695T and BLM-T99A, were studied in stable transfected cells.The nucleoside analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were used to label and track genomic regions of active replication. Cells were initially treated with IdU for 15 minutes. Subsequently, either 4 mM of hydroxyurea (HU) or 30 µM of aphidicolin (Aph) were used to induce replication stress for 4 hours, after which cells were treated with CldU for 85 minutes to assess replication recovery. Cells were mounted on microscope slides, lysed to isolate chromosome fibers, and then fixed on the slides. Sites of IdU and CldU incorporation into the growing DNA strand were then detected by immunostaining the isolated chromosome fibers with analog-specific, fluorescent-coupled antibodies. Replication activity was visualized via microscopy and measured.
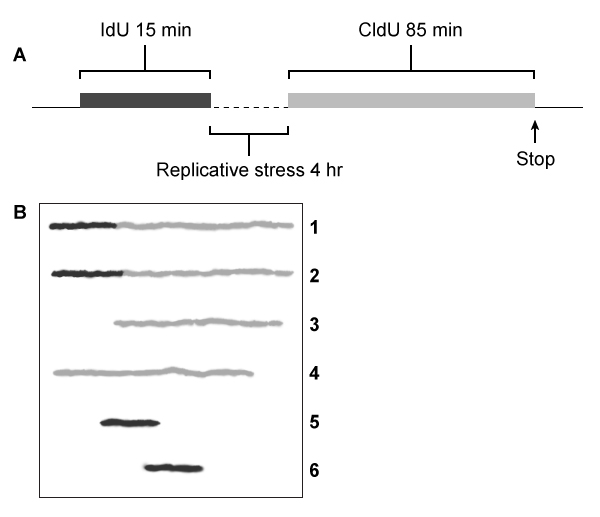 Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
Figure 1 (A) Experimental protocol and (B) fluorescent imaging of a microscope slide showing incorporation of IdU (black lines) and CldU (gray lines) into DNA fibers isolated from a group of BLM+ and BLM− cells, with single fibers depicted in each row
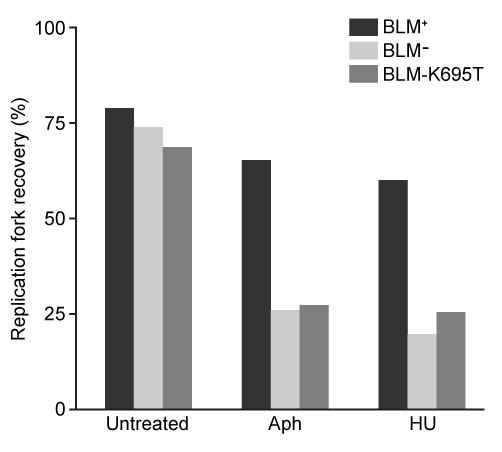 Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
Figure 2 Replication fork recovery in cells expressing BLM+, BLM−, and the mutation BLM-K695T (antagonistic to wild-type)
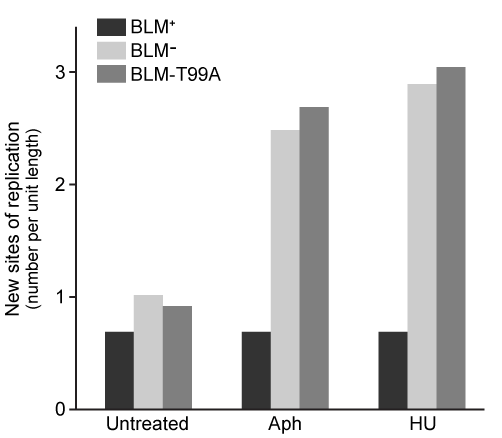 Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Figure 3 New sites of replication in cells expressing BLM+, BLM−, and the mutation BLM-T99A (T99 phosphorylated after replication arrest)
Adapted from Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24(3) :1279-91.
-After the 85-minute period, deoxyribonucleoside triphosphates (dNTPs) are added and DNA synthesis is allowed to continue for 20 minutes. Which of the following processes is most likely to occur during the new replication period? The structure of CldU is shown below. 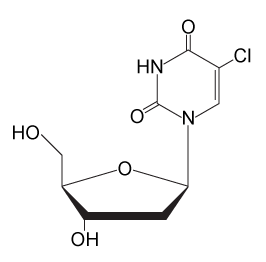
A) CldU and a dNTP molecule would react endergonically to release a pyrophosphate molecule.
B) CldU and a dNTP molecule would react exergonically to form a covalent phosphodiester bond.
C) The 3′ OH group from the last nucleotide of the growing strand would attack the 5′ PO4 group of an incoming dNTP.
D) The 5′ PO4 group from the last nucleotide of the growing strand would attack the 3′ OH group of an incoming dNTP.
Correct Answer:
Verified
Q17: Passage
At the blood-brain barrier, the cerebral spinal
Q18: Passage
Quantitative fluorescence recovery after photobleaching (FRAP) is
Q19: Passage
Quantitative fluorescence recovery after photobleaching (FRAP) is
Q20: Passage
The lumen of the human gut is
Q21: Passage
Lipopolysaccharides (LPS) found in gram-negative bacterial cells
Q23: Passage
Hematological anomalies have been observed in patients
Q24: Passage
Hematological anomalies have been observed in patients
Q25: Passage
The Bloom syndrome helicase (BLM) transcript is
Q26: Passage
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a
Q27: Passage
The Bloom syndrome helicase (BLM) transcript is
Unlock this Answer For Free Now!
View this answer and more for free by performing one of the following actions

Scan the QR code to install the App and get 2 free unlocks

Unlock quizzes for free by uploading documents