Passage
A limited number of cellular functions exist for D-amino acids, such as the structural role of D-alanine and D-glutamate in bacterial cell walls. Because genetically encoded amino acids are synthesized in the L form, production of D-amino acids depends on enzymes called racemases. With the exception of cysteine, conversion of an L-amino acid to a D-amino acid corresponds to the conversion of an S stereoisomer to an R stereoisomer. The mechanism of conversion requires the formation of a high-energy carbanion intermediate. Although pyridoxal phosphate (PLP) -dependent amino acid racemases such as glutamate racemase use PLP as a coenzyme to stabilize this intermediate, the reaction catalyzed by PLP-independent racemases such as alanine racemase proceeds through the enolate intermediate shown in Figure 1.
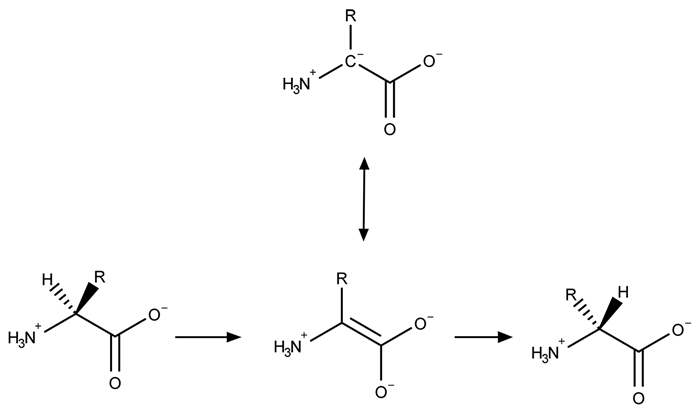 Figure 1 PLP-independent amino acid racemase reactionThe first step in most PLP-dependent reactions is the condensation of an aldehyde group on the coenzyme PLP. This forms a Schiff base linking PLP to a lysine side chain on the enzyme's active site. The lysine is then substituted with the amino acid to be converted from L to D configuration (Figure 2) .
Figure 1 PLP-independent amino acid racemase reactionThe first step in most PLP-dependent reactions is the condensation of an aldehyde group on the coenzyme PLP. This forms a Schiff base linking PLP to a lysine side chain on the enzyme's active site. The lysine is then substituted with the amino acid to be converted from L to D configuration (Figure 2) .
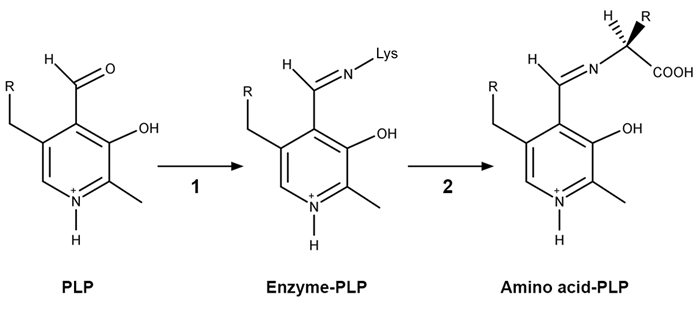 Figure 2 Formation of an amino acid-PLP adductThe next step in PLP-dependent reactions is the removal of the amino acid's α-hydrogen by a base in the enzyme's active site. A quinonoid intermediate and two additional intermediates are formed, as shown in Figure 3. The final step includes the rebonding of PLP to the enzyme's active site and the release of the D-amino acid.
Figure 2 Formation of an amino acid-PLP adductThe next step in PLP-dependent reactions is the removal of the amino acid's α-hydrogen by a base in the enzyme's active site. A quinonoid intermediate and two additional intermediates are formed, as shown in Figure 3. The final step includes the rebonding of PLP to the enzyme's active site and the release of the D-amino acid.
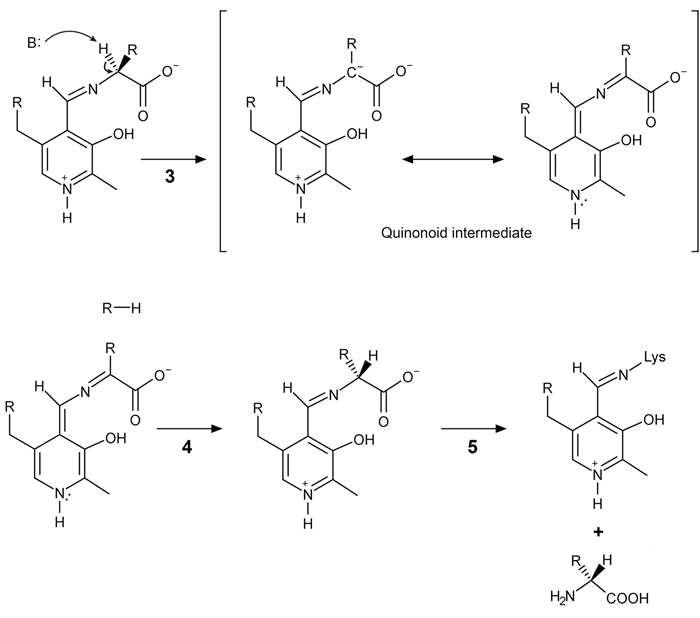 Figure 3 Intermediates of a PLP-dependent amino acid racemase-catalyzed reactionOther PLP-dependent enzymes participate in the decarboxylation of amino acids via a similar mechanism. Such enzymes break the bond between the carboxylate carbon and α-carbon in an amino acid by forming a quinonoid intermediate, which helps stabilize the carbanion intermediate of the reaction.
Figure 3 Intermediates of a PLP-dependent amino acid racemase-catalyzed reactionOther PLP-dependent enzymes participate in the decarboxylation of amino acids via a similar mechanism. Such enzymes break the bond between the carboxylate carbon and α-carbon in an amino acid by forming a quinonoid intermediate, which helps stabilize the carbanion intermediate of the reaction.
Adapted from Cava F, Lam H, De pedro MA, Waldor MK. Emerging knowledge of regulatory roles of D-amino acids in bacteria. Cell Mol Life Sci. 2011.
-The Schiff base shown in Figure 2 involves the formation of what nitrogen-containing functional group?
A) Amide
B) Enamine
C) Imide
D) Imine
Correct Answer:
Verified
Q6: Passage
A limited number of cellular functions exist
Q7: Passage
The drug paracetamol, also known as acetaminophen,
Q8: Passage
Hemophilia B is a blood clotting disorder
Q9: Passage
Hemophilia B is a blood clotting disorder
Q10: Passage
The drug paracetamol, also known as acetaminophen,
Q12: Passage
The drug paracetamol, also known as acetaminophen,
Q13: Passage
High blood pressure, or hypertension, is a
Q14: Passage
A limited number of cellular functions exist
Q15: Passage
A limited number of cellular functions exist
Q16: Passage
The drug paracetamol, also known as acetaminophen,
Unlock this Answer For Free Now!
View this answer and more for free by performing one of the following actions

Scan the QR code to install the App and get 2 free unlocks

Unlock quizzes for free by uploading documents