Passage
The HIV-derived transactivator of transcription (Tat) is a short peptide with the sequence RKKRRQRRRPPQ. When paired with liposomes (ie, synthetic vesicles) composed of cationic lipids, Tat can transport large molecules across cell membranes and has been studied as a vector for gene therapy. However, Tat delivers cargo to cells with relatively low efficiency. Researchers hypothesized that a Tat homodimer could yield stronger DNA binding than the monomer, which may improve the efficiency of DNA delivery.To study this hypothesis, Tat and two variant peptides (Variants 1 and 2) were produced by solid-phase peptide synthesis, in which the growing peptide chain is immobilized on polystyrene beads. Unlike biological systems, solid-phase synthesis assembles peptides from the C-terminus to the N-terminus, as shown in Figure 1.
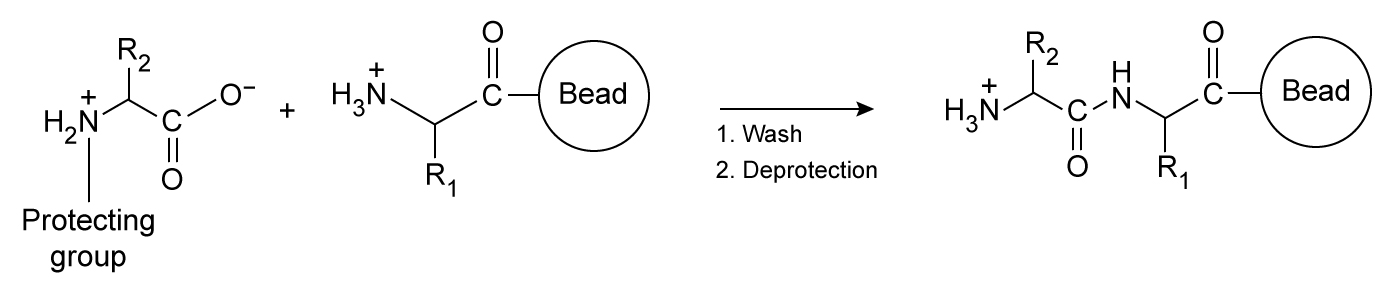 Figure 1 Simplified schematic of one round of solid-phase peptide synthesis. The protecting group is removed during deprotection. The process is repeated until the peptide is complete.Variants 1 and 2 contained a C-terminal and an N-terminal cysteine residue, respectively. Each variant was allowed to dimerize by disulfide bond formation. Peptide-DNA binding interactions were then studied by mixing varying amounts of each peptide with plasmid DNA encoding luciferase protein (pLuc) . The resulting peptide-DNA complexes were run on agarose gels, with results displayed in Figure 2.
Figure 1 Simplified schematic of one round of solid-phase peptide synthesis. The protecting group is removed during deprotection. The process is repeated until the peptide is complete.Variants 1 and 2 contained a C-terminal and an N-terminal cysteine residue, respectively. Each variant was allowed to dimerize by disulfide bond formation. Peptide-DNA binding interactions were then studied by mixing varying amounts of each peptide with plasmid DNA encoding luciferase protein (pLuc) . The resulting peptide-DNA complexes were run on agarose gels, with results displayed in Figure 2.
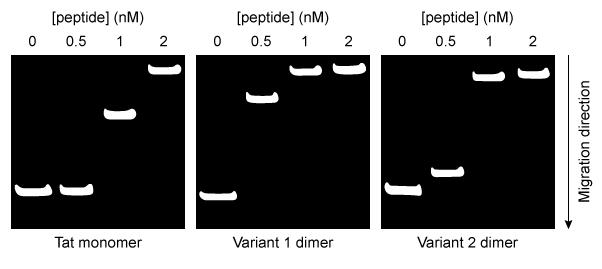 Figure 2 Agarose gel results for pLuc plasmid exposed to varying amounts of each peptideEqual concentrations of each type of peptide-DNA complex were then incubated with cationic liposomes and exposed to cultured cells for 24 hours. The cells were then assessed for luciferase activity. Greater luciferase activity indicated more efficient delivery of the pLuc plasmid. The results are shown in Figure 3.
Figure 2 Agarose gel results for pLuc plasmid exposed to varying amounts of each peptideEqual concentrations of each type of peptide-DNA complex were then incubated with cationic liposomes and exposed to cultured cells for 24 hours. The cells were then assessed for luciferase activity. Greater luciferase activity indicated more efficient delivery of the pLuc plasmid. The results are shown in Figure 3.
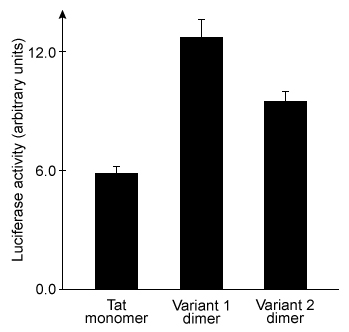 Figure 3 Plasmid delivery efficiency of three peptides, as measured by induced luciferase activity
Figure 3 Plasmid delivery efficiency of three peptides, as measured by induced luciferase activity
Adapted from Lee S-J, Yoon S-H, Doh K-O. Enhancement of gene delivery using novel homodimeric tat peptide formed by disulfide bond. J Microbiol Biotechnol. 2011;21(8) :802-807.
-Do the data in Figure 2 and Figure 3 support the hypothesis that stronger peptide-DNA interactions yield more efficient plasmid delivery?
A) Yes; the order of binding strength shown in Figure 2 is the same as the order of delivery efficiency shown in Figure 3.
B) No; the Variant 2 dimer shows the weakest DNA binding but is more efficient than the Tat monomer at cargo delivery.
C) No; the Tat monomer shows the strongest DNA binding but the least efficient plasmid delivery.
D) No; the Variant 2 dimer binds DNA with greater strength than the Variant 1 dimer, but Variant 1 has the most efficient plasmid delivery.
Correct Answer:
Verified
Q296: Passage
Glycolysis and gluconeogenesis are tightly regulated, opposing
Q297: Certain RNA molecules can fold into structures
Q298: An enzyme generates a Lineweaver-Burk plot with
Q299: Passage
Glycolysis and gluconeogenesis are tightly regulated, opposing
Q300: Passage
Destabilase, an enzyme found in the saliva
Q302: Passage
The HIV-derived transactivator of transcription (Tat) is
Q303: The electron transport chain results in the
Q304: Passage
The HIV-derived transactivator of transcription (Tat) is
Q305: Passage
The HIV-derived transactivator of transcription (Tat) is
Q306: Passage
The HIV-derived transactivator of transcription (Tat) is
Unlock this Answer For Free Now!
View this answer and more for free by performing one of the following actions

Scan the QR code to install the App and get 2 free unlocks

Unlock quizzes for free by uploading documents