Passage
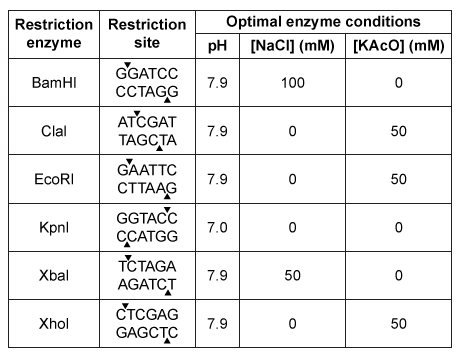
In the central nervous system, myelin produced by oligodendrocytes (glial cells) functions as an insulating sheath surrounding certain nerve fibers. To identify candidate genes involved in the myelination process, researchers collected oligodendrocytes from zebrafish and purified the messenger RNA (mRNA) transcripts expressed by these glial cells. The mRNA was converted to complementary DNA (cDNA) by reverse transcriptase, and the cDNA was fluorescently labeled and assessed for hybridization on a microarray. The expression of mRNA in other zebrafish cells was similarly measured.Transcripts detected in zebrafish oligodendrocytes at levels greater than 3 times those found in other cells were selected as candidate myelination genes. Some of the detected genes, including plp1a, Sox10, and mbp, were previously known to be specific to oligodendrocytes, validating the procedure. The cDNA of newly identified candidates was amplified by PCR, and the ends were digested with the restriction enzymes EcoRI and XhoI. Genes were then ligated into the multiple cloning site (MCS) of the pSKII vector, shown in Figures 1 and 2, which had also been digested by EcoRI and XhoI.
 Figure 1 Overview of the pSKII vector (plasmid)
Figure 1 Overview of the pSKII vector (plasmid)
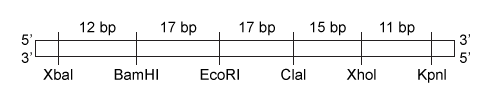 Figure 2 The pSKII MCS, showing the restriction sites of several restriction enzymesTable 1 shows the functional parameters of several restriction enzymes.
Figure 2 The pSKII MCS, showing the restriction sites of several restriction enzymesTable 1 shows the functional parameters of several restriction enzymes.
 To confirm that the candidate genes originated in oligodendrocytes, complementary RNA (cRNA) strands were synthesized from the cloned plasmids and hybridized to fixed zebrafish sections. One of the genes that localized to oligodendrocytes, known as cldnk, produces a 915 base pair mRNA transcript from two exons. The cldnk gene is involved in tight junction formation and may be required for myelin sheath development around axons.
To confirm that the candidate genes originated in oligodendrocytes, complementary RNA (cRNA) strands were synthesized from the cloned plasmids and hybridized to fixed zebrafish sections. One of the genes that localized to oligodendrocytes, known as cldnk, produces a 915 base pair mRNA transcript from two exons. The cldnk gene is involved in tight junction formation and may be required for myelin sheath development around axons.
-What observation could have led researchers to conclude that cldnk is expressed in oligodendrocytes?
A) A probe made of cldnk mRNA hybridized to RNA in cells that express Sox10.
B) An RNA probe that was complementary to cldnk mRNA hybridized in cells that express plp1a.
C) A cldnk cDNA probe obtained from oligodendrocytes bound to a microarray with higher affinity than the same probe obtained from other cells.
D) A cDNA probe corresponding to cldnk hybridized to the nuclei of oligodendrocytes.
Correct Answer:
Verified
Q278: During a nutrition study, a participant consumes
Q279: Renal studies in mice have revealed that
Q280: Passage
Trypanosoma brucei is a eukaryotic parasite that
Q281: Dehydration caused by frequent loose stools is
Q282: Passage
In the central nervous system, myelin produced
Q284: A physician finds that a patient experiences
Q285: Passage
Within the Actinopterygii (bony fish) lineage, marine
Q286: Passage
Within the Actinopterygii (bony fish) lineage, marine
Q287: Passage
In the central nervous system, myelin produced
Q288: Passage
In the central nervous system, myelin produced
Unlock this Answer For Free Now!
View this answer and more for free by performing one of the following actions

Scan the QR code to install the App and get 2 free unlocks

Unlock quizzes for free by uploading documents