The specificity of nucleic acid hybridization is remarkable. Even a single mismatch between a probe and a target sequence can be detected depending on hybridization conditions. This sensitivity can be employed in applications such as SNP haplotyping. Consider a probe that is complementary to a 20-nucleotide-long genomic region including a single polymorphic nucleotide. The probe is perfectly complementary to the more common allele, but has one mismatch in the case of the minor allele. The temperature-dependent hybridization of the probe to the single-strand genomic fragment is schematically presented in the following graph. To detect hybridization, a fluorescent dye is added to the reaction. The dye fluoresces strongly only when intercalated between the bases of double-stranded DNA, and is therefore used to quantify double-strand formation or dissociation. Which "melting curve" (1 or 2) in the graph do you think corresponds to the more common SNP allele? Which curve shows a higher melting temperature? 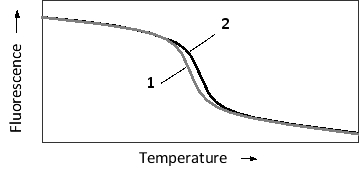
A) 1; 1
B) 1; 2
C) 2; 1
D) 2; 2
Correct Answer:
Verified
Q24: In an unfolded (random coil) protein, amino
Q25: Indicate whether each of the following manipulations
Q26: Your friend has cut a 3000-nucleotide-pair circular
Q27: You have carried out a genetic screen
Q28: You have used a variation of fluorescence
Q30: The binding of protein A to two
Q31: Indicate whether each of the following descriptions
Q32: Each cycle of PCR doubles the amount
Q33: In sequence alignments such as those generated
Q34: Indicate whether each of the following descriptions
Unlock this Answer For Free Now!
View this answer and more for free by performing one of the following actions

Scan the QR code to install the App and get 2 free unlocks

Unlock quizzes for free by uploading documents