Passage
The primary structure of a protein is a series of amino acids that are connected in a long chain via C−N peptide bonds (Figure 1) .
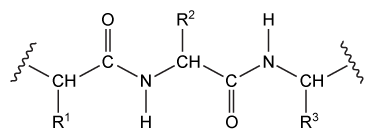 Figure 1 Peptide bonds within a protein segment (Note: Rn = amino acid side chains) Protein chain folding is guided by multiple intramolecular interactions that can occur between two peptide bonds, between a peptide bond and a side chain, or between two side chains, as shown in the protein in Figure 2. Although these interactions are intramolecular because they are within the same molecule, they act effectively like intermolecular interactions because proteins are such large molecules.
Figure 1 Peptide bonds within a protein segment (Note: Rn = amino acid side chains) Protein chain folding is guided by multiple intramolecular interactions that can occur between two peptide bonds, between a peptide bond and a side chain, or between two side chains, as shown in the protein in Figure 2. Although these interactions are intramolecular because they are within the same molecule, they act effectively like intermolecular interactions because proteins are such large molecules.
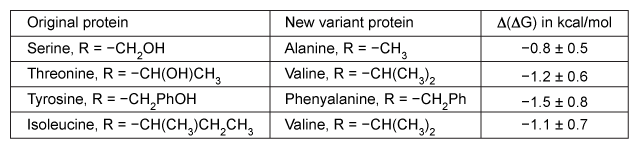
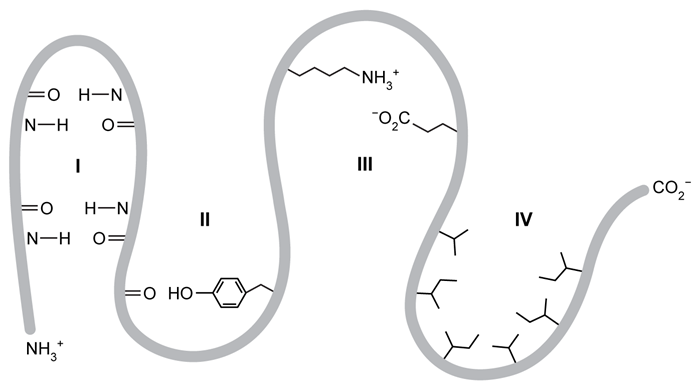 Figure 2 Intramolecular interactions between the peptide chain and amino acid side chains within a hypothetical proteinBecause intramolecular interactions play a significant role in maintaining the three-dimensional folded structure of a protein, having a greater number of interactions or stronger interactions will result in increased protein stability. The energy required to break these interactions within a protein is related to free energy, ΔG. The greater the ΔG value, the more stable the protein.To find the overall contribution of particular intramolecular interactions to protein stability, researchers made several protein variants in which one amino acid is replaced by another to change the noncovalent interactions between the side chains. The difference in free energy between the variant and the original protein, Δ(ΔG) , indicates the change in overall stability of the protein. A negative Δ(ΔG) means the variant protein is less stable than the original.Table 1 shows the results of the amino acid substitution experiments for a protein with the approximate energy difference when each amino acid is changed.Table 1 Change in Free Energy Based on Changing Intramolecular Interactions Caused by Amino Acid Substitutions
Figure 2 Intramolecular interactions between the peptide chain and amino acid side chains within a hypothetical proteinBecause intramolecular interactions play a significant role in maintaining the three-dimensional folded structure of a protein, having a greater number of interactions or stronger interactions will result in increased protein stability. The energy required to break these interactions within a protein is related to free energy, ΔG. The greater the ΔG value, the more stable the protein.To find the overall contribution of particular intramolecular interactions to protein stability, researchers made several protein variants in which one amino acid is replaced by another to change the noncovalent interactions between the side chains. The difference in free energy between the variant and the original protein, Δ(ΔG) , indicates the change in overall stability of the protein. A negative Δ(ΔG) means the variant protein is less stable than the original.Table 1 shows the results of the amino acid substitution experiments for a protein with the approximate energy difference when each amino acid is changed.Table 1 Change in Free Energy Based on Changing Intramolecular Interactions Caused by Amino Acid Substitutions
 Adapted from: C. N. Pace, J. M. Scholtz, G. R. Grimsley, "Forces stabilizing proteins." FEBS Lett. ©2014 Wiley.
Adapted from: C. N. Pace, J. M. Scholtz, G. R. Grimsley, "Forces stabilizing proteins." FEBS Lett. ©2014 Wiley.
-In the protein shown in Figure 2, the folding of region I is predominantly stabilized by:
A) salt-bridge interactions between amino acid side chains.
B) covalent bonding between peptide segments.
C) hydrophobic interactions between amino acid side chains.
D) hydrogen bonding between peptide segments.
Correct Answer:
Verified
Q161: Passage
The primary structure of a protein is
Q162: Some test strips for blood glucose meters
Q163: What are the correct formal charges for
Q164: If MgCO3 undergoes a double replacement reaction
Q165: A 0.040 M HCl solution is measured
Q167: The electrolysis of aqueous silver nitrate involves
Q168: Oxalate anions, C2O42−, added to a solution
Q169: Osmolarity, the number of moles of osmotically
Q170: If each of the following is an
Q171: Which of the following values gives the
Unlock this Answer For Free Now!
View this answer and more for free by performing one of the following actions

Scan the QR code to install the App and get 2 free unlocks

Unlock quizzes for free by uploading documents