You have performed a chromosome conformation capture (3C) experiment to study chromatin looping at a mouse gene cluster that contains genes A, B, and C, as well as a regulatory region R. In this experiment, you performed in situ chemical cross-linking of chromatin, followed by cleavage of DNA in the nuclear extract with a restriction enzyme, intramolecular ligation, and cross-link removal. Finally, a polymerase chain reaction (PCR) was carried out using a forward primer that hybridizes to a region in the active B gene, and one of several reverse primers, each of which hybridizes to a different location in the locus. The amounts of the PCR products were quantified and normalized to represent the relative cross-linking efficiency in each analyzed sample. You have plotted the results in the graph below. The same experiment has been done on two tissue samples: fetal liver (represented by red lines) and fetal muscle (blue lines).
Interaction of the A, B, and C genes with the regulatory R region is known to enhance expression of these genes. Indicate true (T) and false (F) statements below based on the results. Your answer would be a four-letter string composed of letters T and F only, e.g. TTTF.
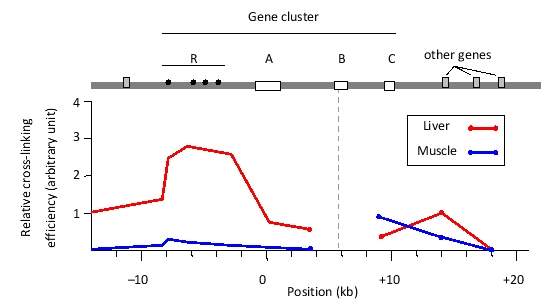
( ) The B gene would be predicted to have higher expression in the fetal liver compared to the fetal muscle tissue.
( ) Interaction between the R region and the B gene involves the A gene looping out.
( ) Interaction between the R region and the B gene involves the C gene looping out.
( ) In fetal muscle, the B gene definitely does not engage in looping interactions with any other elements in the cluster.
Correct Answer:
Verified
The 3C data show high relative cross-l...
View Answer
Unlock this answer now
Get Access to more Verified Answers free of charge
Q20: Which of the following features of DNA
Q21: The centromeric regions in the fission yeast
Q22: Polytene chromosomes are useful for studying chromatin
Q23: For each of the following classifications, indicate
Q24: Nucleosomes that are positioned like beads on
Q26: Indicate whether each of the following descriptions
Q27: A chromatin "reader complex" …
A) is always
Q28: Imagine a chromosome translocation event that brings
Q29: Five major types of chromatin were identified
Q30: In human cells, the alpha satellite DNA
Unlock this Answer For Free Now!
View this answer and more for free by performing one of the following actions

Scan the QR code to install the App and get 2 free unlocks

Unlock quizzes for free by uploading documents